Andrew Carroll
@acarroll.bsky.social
2.6K followers
200 following
18 posts
Product lead Genomics Google Research
Posts
Media
Videos
Starter Packs
Andrew Carroll
@acarroll.bsky.social
· May 13
Andrew Carroll
@acarroll.bsky.social
· May 13

Release DeepSomatic 1.9.0 · google/deepsomatic
DeepSomatic:
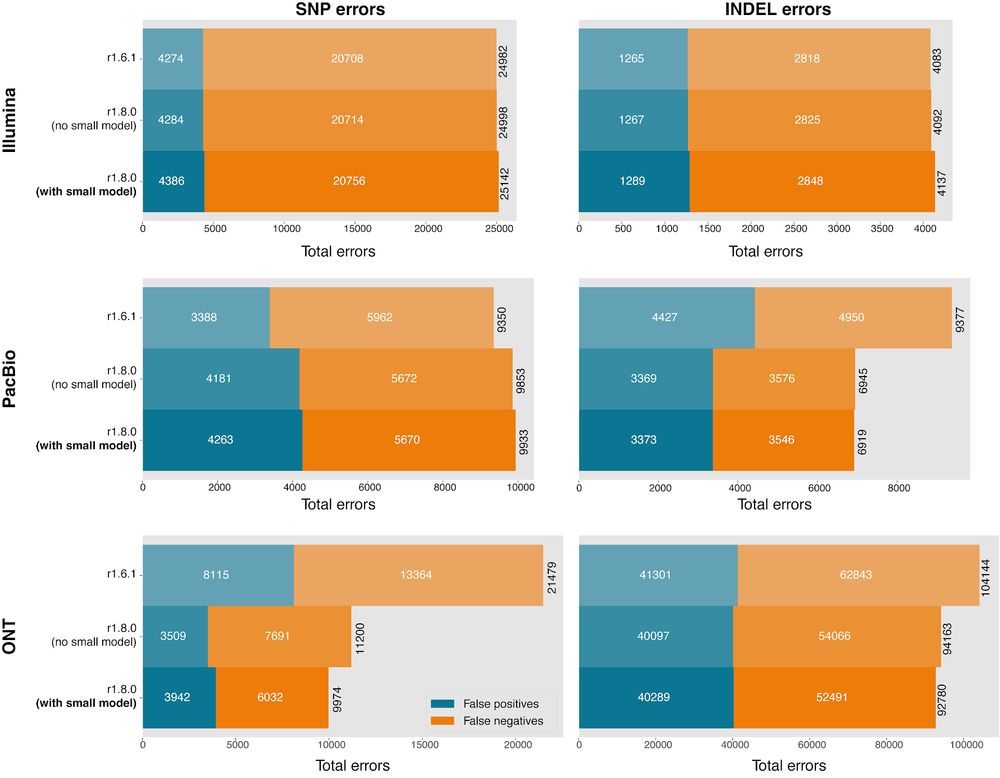
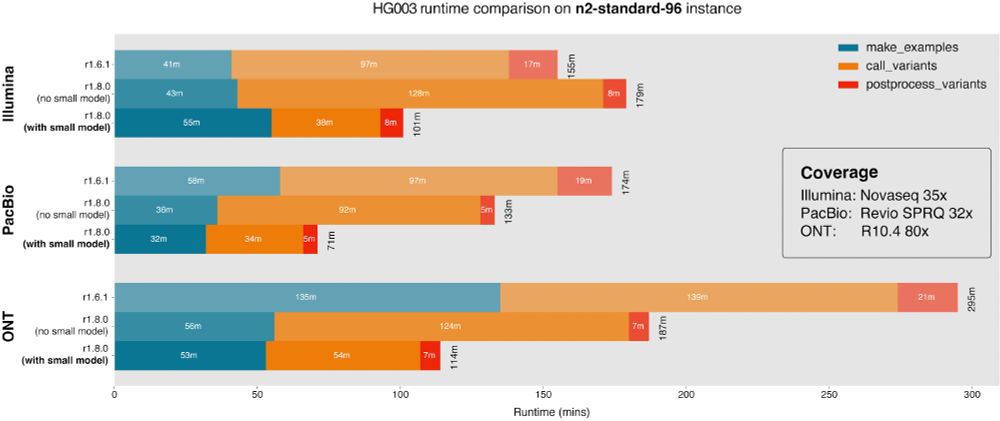
In this release, we are introducing FFPE_WGS_TUMOR_ONLY and FFPE_WES_TUMOR_ONLY models.
The WGS and WGS_TUMOR_ONLY models have been retrained with all datasets described in the manusc...
github.com
Andrew Carroll
@acarroll.bsky.social
· May 13

Release DeepVariant 1.9.0 · google/deepvariant
DeepVariant:
In this version we have updated our training scheme for the HG002 sample with the newly released HG002-T2T truth set which improves accuracy against that truth set.
Our labeling metho...
github.com
Reposted by Andrew Carroll
Andrew Carroll
@acarroll.bsky.social
· Dec 6
Andrew Carroll
@acarroll.bsky.social
· Dec 6
Andrew Carroll
@acarroll.bsky.social
· Dec 5
Andrew Carroll
@acarroll.bsky.social
· Oct 26
Andrew Carroll
@acarroll.bsky.social
· Oct 26
Andrew Carroll
@acarroll.bsky.social
· Oct 26
Andrew Carroll
@acarroll.bsky.social
· Oct 26

Release DeepVariant 1.6.0 · google/deepvariant
Improved support for haploid regions, chrX and chY. Users can specify haploid regions with a flag. Updated case studies show usage and metrics.
Added pangenome workflow (FASTQ-to-VCF mapping with V...
github.com
Reposted by Andrew Carroll
Andrew Carroll
@acarroll.bsky.social
· Oct 24

GitHub - KolmogorovLab/Severus: A tool for somatic structural variant calling using long reads
A tool for somatic structural variant calling using long reads - GitHub - KolmogorovLab/Severus: A tool for somatic structural variant calling using long reads
github.com
Reposted by Andrew Carroll
Alex Rubinsteyn
@alexr.bsky.social
· Sep 14