Adam Cribbs
@adamcribbs.bsky.social
#MRC CDA fellow,
University of Oxford, long-read sequencing| single-cell technology development| multiple myeloma| bone cancer
University of Oxford, long-read sequencing| single-cell technology development| multiple myeloma| bone cancer
work led by
Jianfeng sun,
with fantastic collaborators Shuang Li, Stefan Canzar
Jianfeng sun,
with fantastic collaborators Shuang Li, Stefan Canzar
May 6, 2025 at 9:19 AM
work led by
Jianfeng sun,
with fantastic collaborators Shuang Li, Stefan Canzar
Jianfeng sun,
with fantastic collaborators Shuang Li, Stefan Canzar
UMIche features:
🧠 mclUMI (Markov clustering)
🧬 Homotrimer UMI support (majority vote & set cover)
🧪 scRNA-seq simulation (DeepConvCVAE)
📊 Visual analytics
A step forward in deduplication accuracy & benchmarking.
#UMI #scRNAseq #bioinformatics
🧠 mclUMI (Markov clustering)
🧬 Homotrimer UMI support (majority vote & set cover)
🧪 scRNA-seq simulation (DeepConvCVAE)
📊 Visual analytics
A step forward in deduplication accuracy & benchmarking.
#UMI #scRNAseq #bioinformatics
May 6, 2025 at 9:19 AM
UMIche features:
🧠 mclUMI (Markov clustering)
🧬 Homotrimer UMI support (majority vote & set cover)
🧪 scRNA-seq simulation (DeepConvCVAE)
📊 Visual analytics
A step forward in deduplication accuracy & benchmarking.
#UMI #scRNAseq #bioinformatics
🧠 mclUMI (Markov clustering)
🧬 Homotrimer UMI support (majority vote & set cover)
🧪 scRNA-seq simulation (DeepConvCVAE)
📊 Visual analytics
A step forward in deduplication accuracy & benchmarking.
#UMI #scRNAseq #bioinformatics
With FlowAgent, experimental biologists can efficiently run sophisticated analyses without coding expertise, allowing bioinformaticians to prioritise high-level interpretation and innovation.
March 12, 2025 at 12:07 PM
With FlowAgent, experimental biologists can efficiently run sophisticated analyses without coding expertise, allowing bioinformaticians to prioritise high-level interpretation and innovation.
Benchmarking demonstrated that FlowAgent, powered by GPT-4o, outperforms other models in terms of speed, accuracy, and cost-efficiency, ideal for scalable and diverse bioinformatics analyses.
March 12, 2025 at 12:07 PM
Benchmarking demonstrated that FlowAgent, powered by GPT-4o, outperforms other models in terms of speed, accuracy, and cost-efficiency, ideal for scalable and diverse bioinformatics analyses.
FlowAgent autonomously handles dependency management, tool selection, and real-time parameter optimisation. Intelligent agents continuously monitor computational performance, dataset quality, and software compatibility, providing robust quality control.
March 12, 2025 at 12:07 PM
FlowAgent autonomously handles dependency management, tool selection, and real-time parameter optimisation. Intelligent agents continuously monitor computational performance, dataset quality, and software compatibility, providing robust quality control.
Traditional workflow management systems often require extensive programming skills, limiting usability and adaptability. FlowAgent leverages large language models (LLMs) to translate plain English instructions into structured pipelines, democratising access.
March 12, 2025 at 12:07 PM
Traditional workflow management systems often require extensive programming skills, limiting usability and adaptability. FlowAgent leverages large language models (LLMs) to translate plain English instructions into structured pipelines, democratising access.
Numerous developments and improvements are currently underway following this initial release.
🔗 github.com/EnteloBio/fl...
🔗 github.com/EnteloBio/fl...
GitHub - EnteloBio/flowagent: An advanced multi-agent framework for automating complex bioinformatics workflows.
An advanced multi-agent framework for automating complex bioinformatics workflows. - EnteloBio/flowagent
github.com
March 12, 2025 at 12:07 PM
Numerous developments and improvements are currently underway following this initial release.
🔗 github.com/EnteloBio/fl...
🔗 github.com/EnteloBio/fl...
The anchor-enhanced design demonstrated substantial improvements in data quality, particularly in detecting low-abundance transcripts and improving the transcript coverage.
February 16, 2025 at 10:47 AM
The anchor-enhanced design demonstrated substantial improvements in data quality, particularly in detecting low-abundance transcripts and improving the transcript coverage.
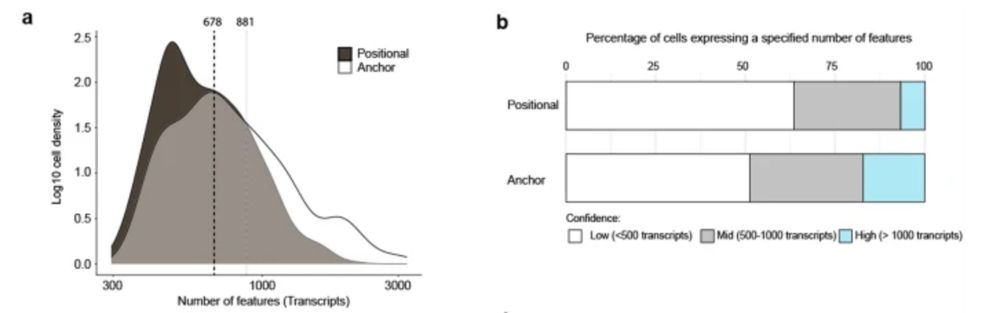
The effects of adding an anchor sequence are a significantly increased number of features per cell over and above not having an anchor (Positional).
February 16, 2025 at 10:47 AM
The effects of adding an anchor sequence are a significantly increased number of features per cell over and above not having an anchor (Positional).
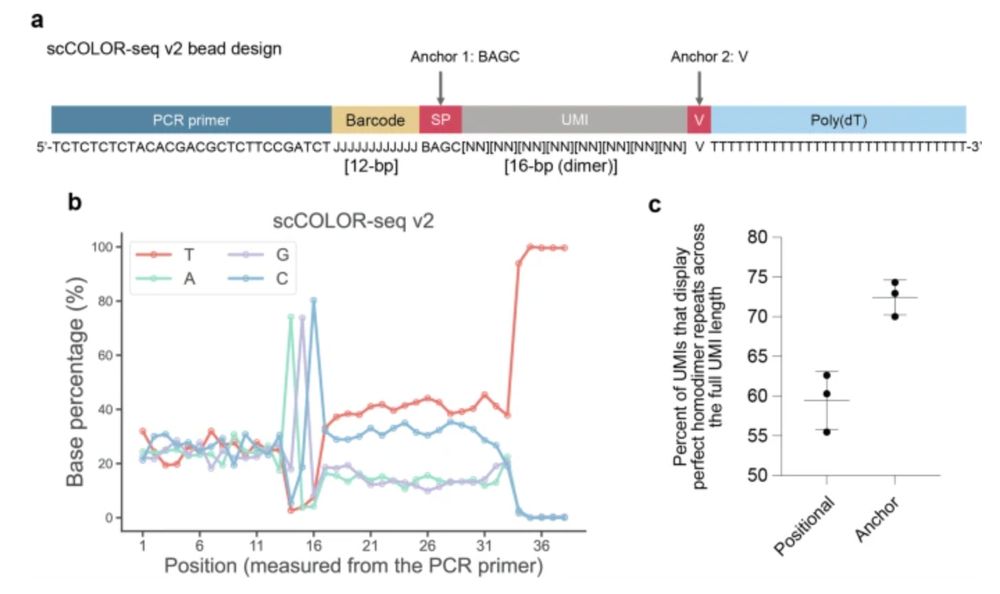
By integrating an anchor sequence before and after the UMI, we observed a significant reduction in technical variability, an increase in UMI accuracy, leading to more accurate gene expression measurements.
February 16, 2025 at 10:47 AM
By integrating an anchor sequence before and after the UMI, we observed a significant reduction in technical variability, an increase in UMI accuracy, leading to more accurate gene expression measurements.
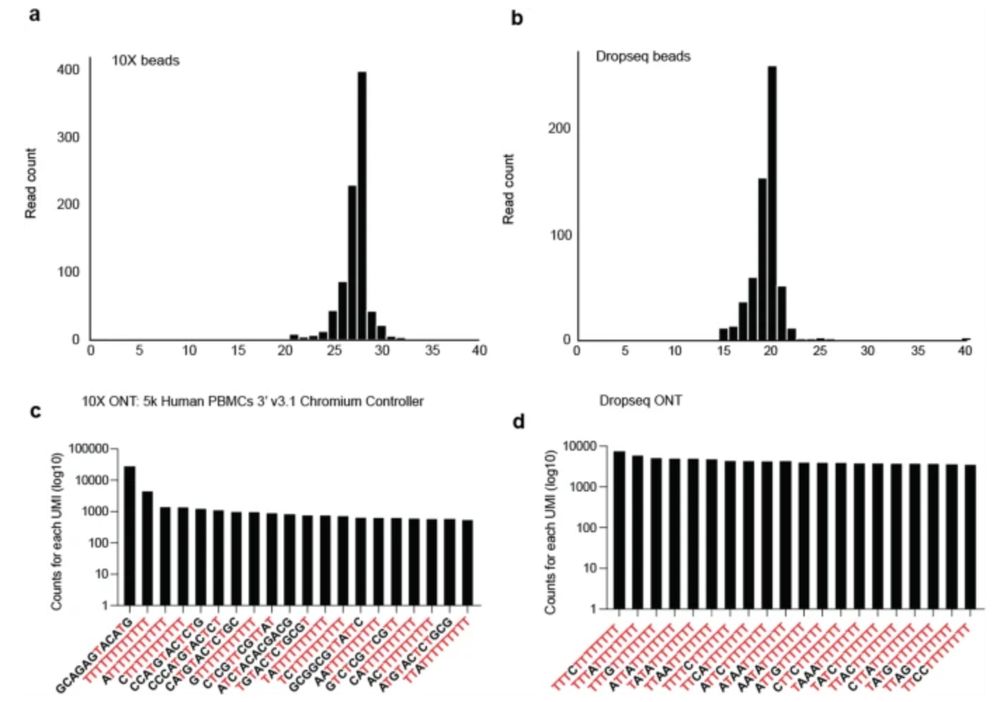
Turns out for droplet based single-cell methods, beads are truncated and this leads to several issues with downstream artefacts. Mainly, there is an overrepresentation of Ts in the UMI that result from truncated bead synthesis.
February 16, 2025 at 10:47 AM
Turns out for droplet based single-cell methods, beads are truncated and this leads to several issues with downstream artefacts. Mainly, there is an overrepresentation of Ts in the UMI that result from truncated bead synthesis.

