Alex Geary
@alextremophile.bsky.social
570 followers
430 following
30 posts
Postdoctoral Bioinformaticial in the Computational Rare Disease Genomics group (Nicky Whiffin). univ. Oxford 🧬💻
Loves Evolution, regulation, cheese and cats. She/Her
Posts
Media
Videos
Starter Packs
Alex Geary
@alextremophile.bsky.social
· Aug 29
Alex Geary
@alextremophile.bsky.social
· Aug 29
Alex Geary
@alextremophile.bsky.social
· Aug 29
Alex Geary
@alextremophile.bsky.social
· Aug 29
Alex Geary
@alextremophile.bsky.social
· Aug 29

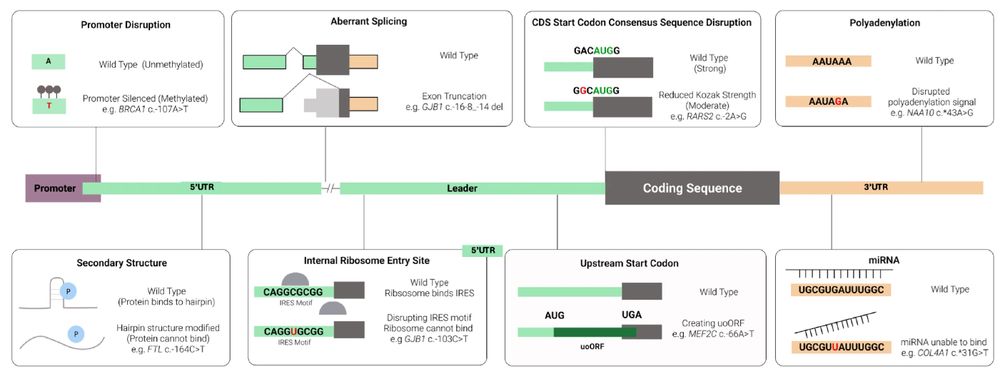
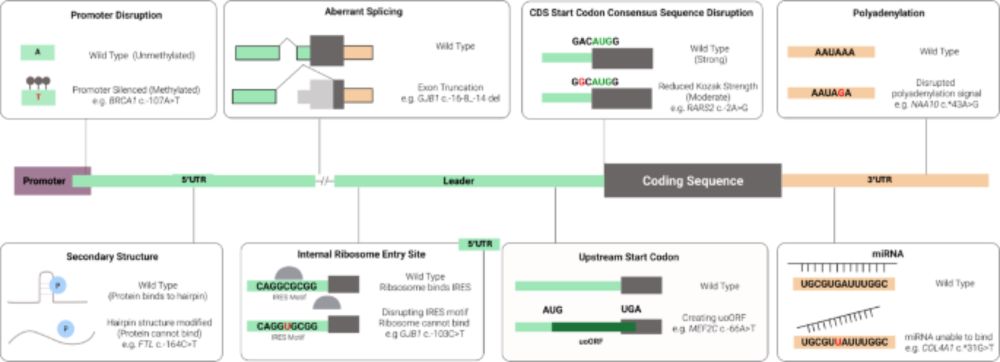
Using SpliceAI to triage splice-altering variants in 7,220 individuals with rare conditions highlights limitations of the precomputed scores
Background: SpliceAI is a deep learning algorithm that predicts whether genetic variants are likely to affect splicing. Precomputed spliceAI predictions for all theoretical SNVs and small indels were ...
www.medrxiv.org
Alex Geary
@alextremophile.bsky.social
· Aug 18
Reposted by Alex Geary
Reposted by Alex Geary
William Shakespeare
@shakespeare.lol
· Apr 25
Reposted by Alex Geary
Dr Zoë Ayres
@zjayres.bsky.social
· Apr 16
Reposted by Alex Geary
Alex Geary
@alextremophile.bsky.social
· Apr 14

a white puppy is laying on the ground with the words `` thank you thank you thank you thank you '' written above it .
ALT: a white puppy is laying on the ground with the words `` thank you thank you thank you thank you '' written above it .
media.tenor.com
Alex Geary
@alextremophile.bsky.social
· Apr 14
Alex Geary
@alextremophile.bsky.social
· Apr 14