
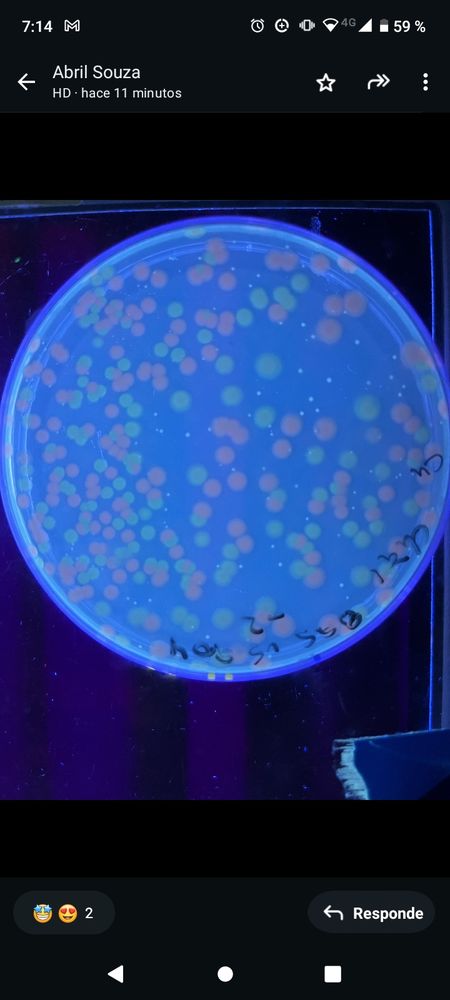
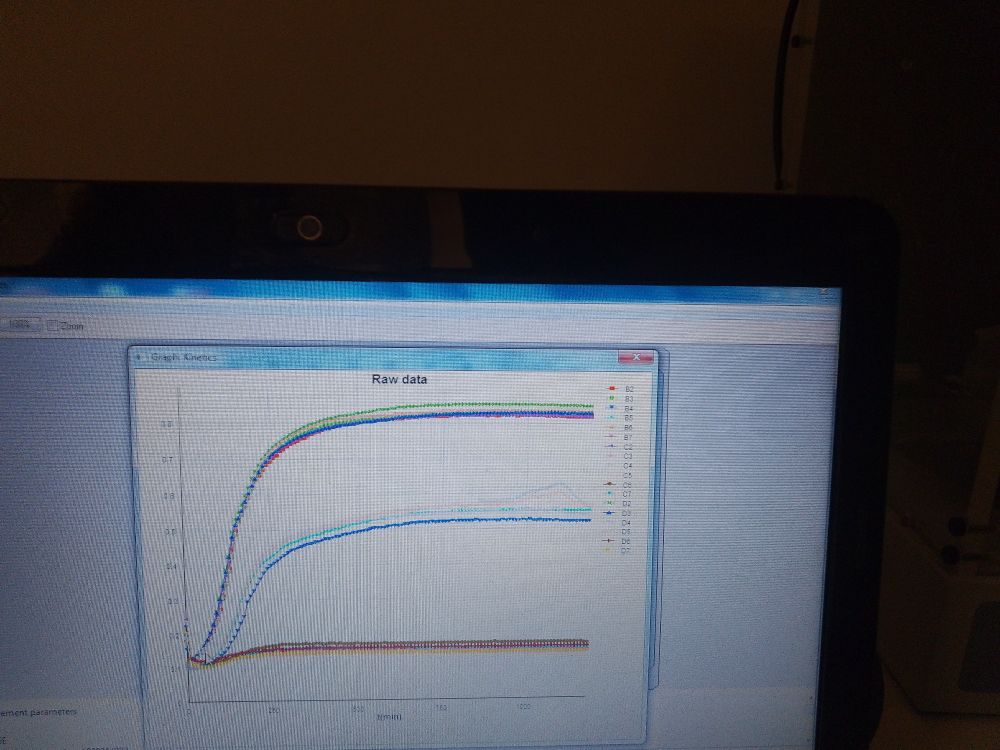
Microbial, growth, chromosome structure & evolution.
Vibrio, Bradyrhizobium (and Brucella) .
❤️🧪🦠Microbiology 🔬🧫❤️
Editor @ Curr. microbiology.
And the many PhD Students and Posdocs implicated!
And the many PhD Students and Posdocs implicated!


asoler1.wixsite.com/iib-geb


asoler1.wixsite.com/iib-geb

