Alisia Fadini
@alisiafadini.bsky.social
310 followers
190 following
20 posts
Researcher. Interested in molecular biophysics using ML + protein structure experiments.
Posts
Media
Videos
Starter Packs
Pinned
Alisia Fadini
@alisiafadini.bsky.social
· Mar 17
Alisia Fadini
@alisiafadini.bsky.social
· Mar 17
Alisia Fadini
@alisiafadini.bsky.social
· Mar 17
Alisia Fadini
@alisiafadini.bsky.social
· Feb 27
Alisia Fadini
@alisiafadini.bsky.social
· Feb 24
Alisia Fadini
@alisiafadini.bsky.social
· Feb 24
Alisia Fadini
@alisiafadini.bsky.social
· Feb 24

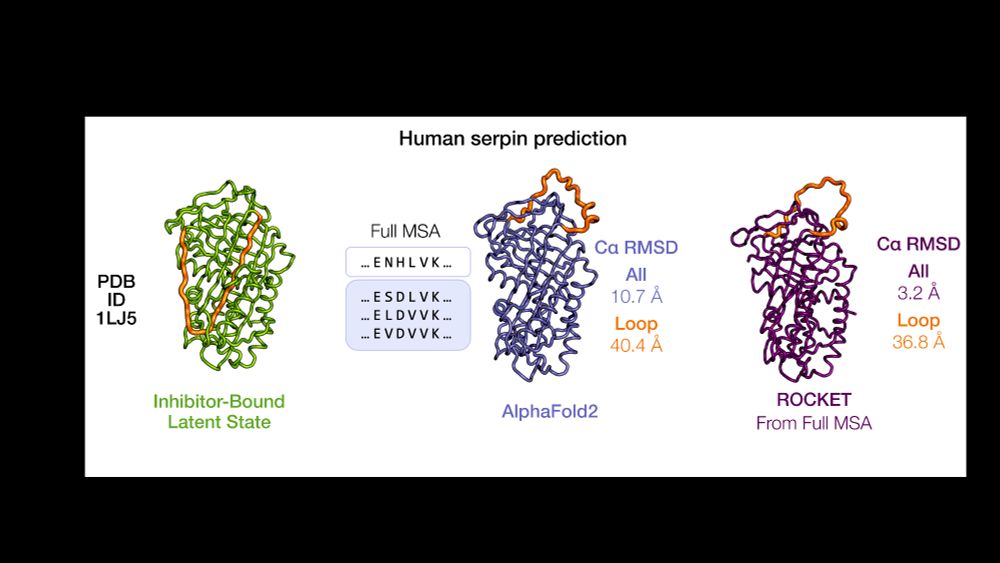
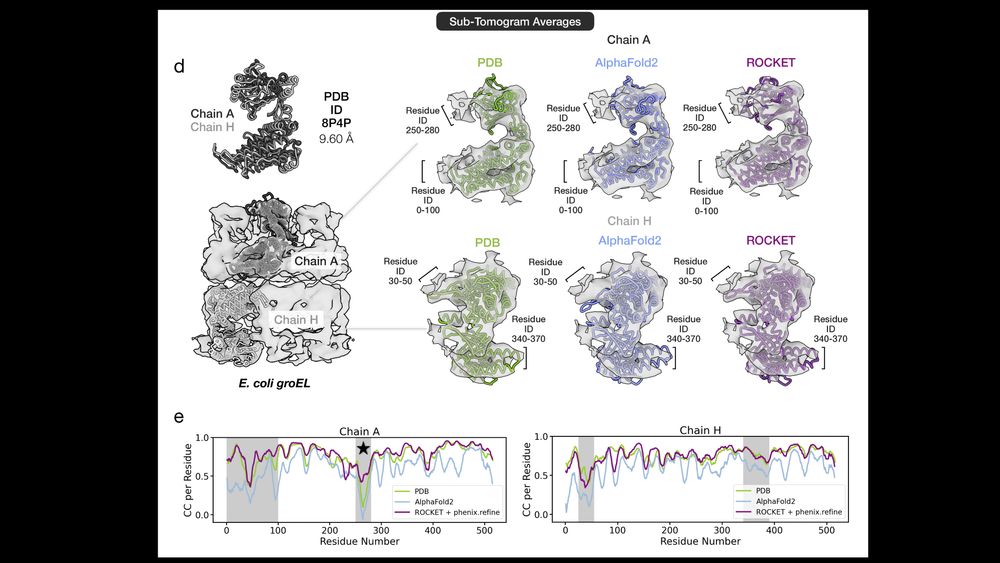
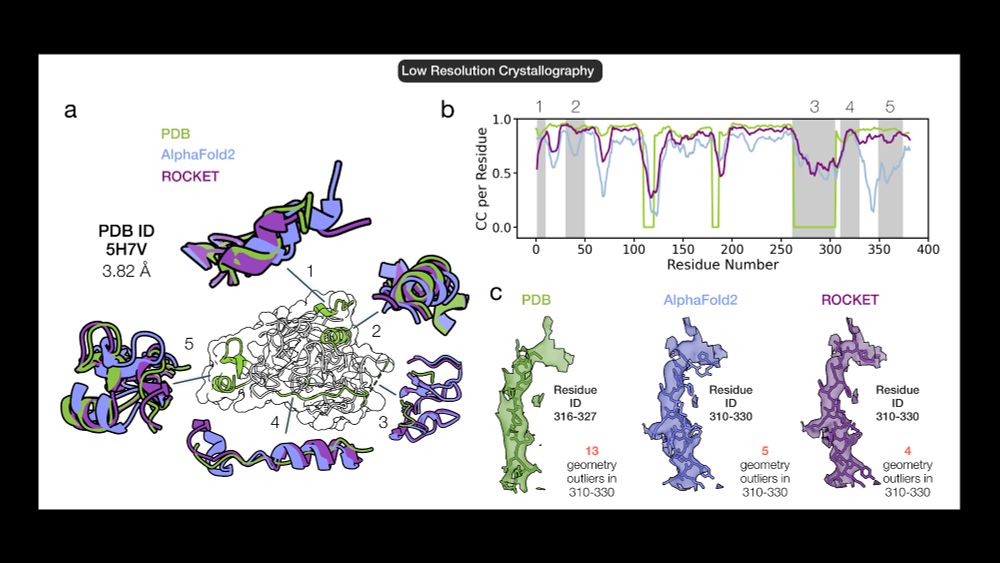
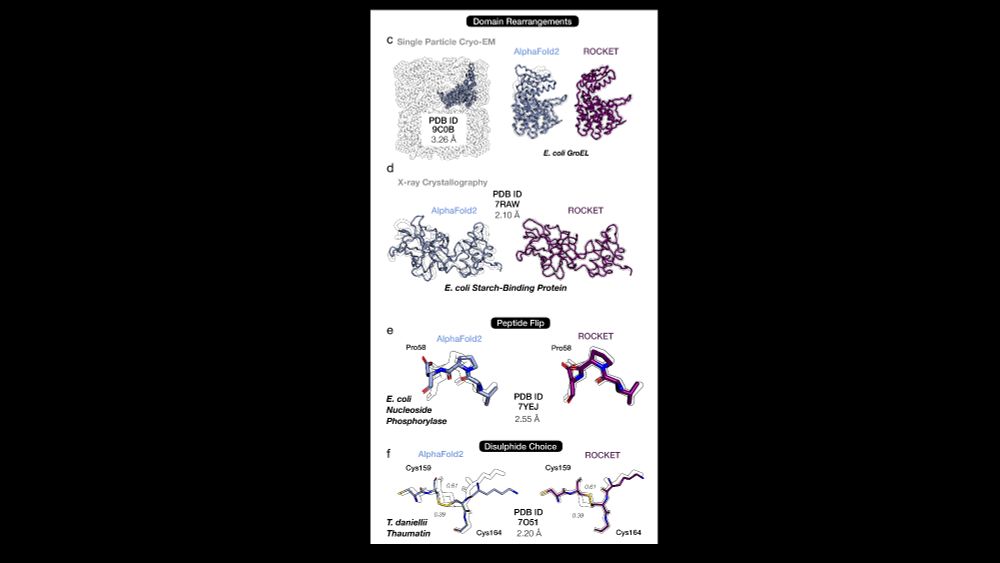
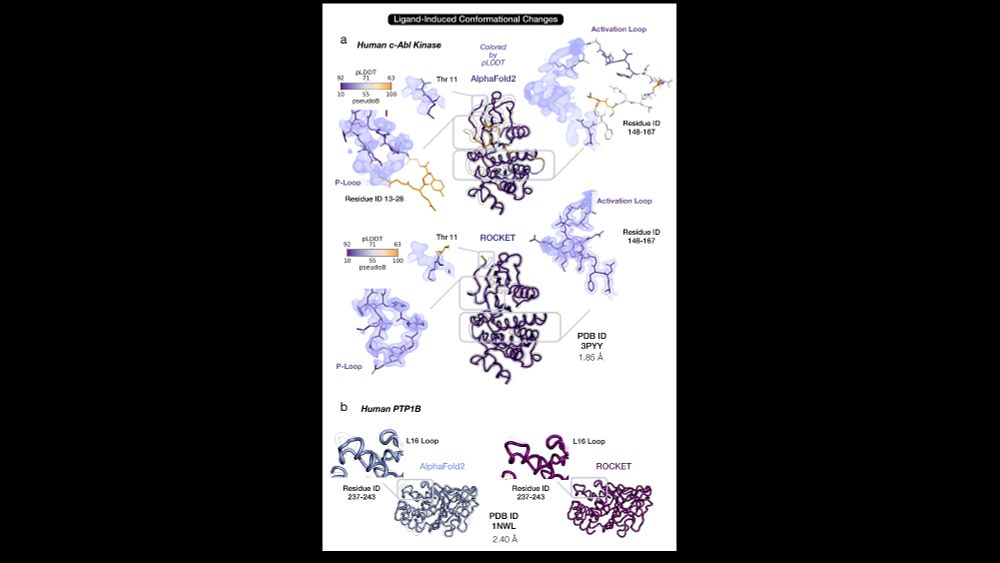
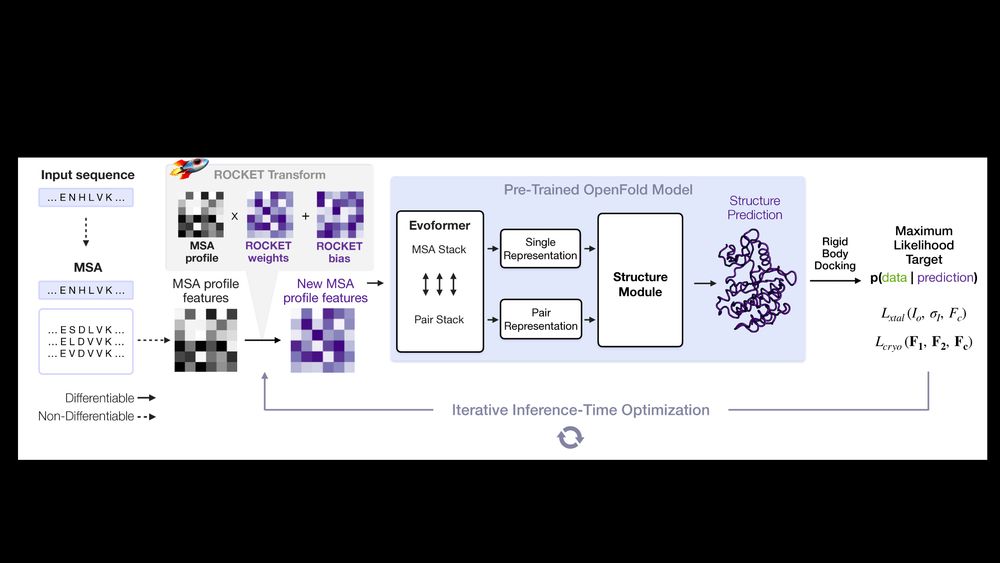
AlphaFold as a Prior: Experimental Structure Determination Conditioned on a Pretrained Neural Network
Advances in machine learning have transformed structural biology, enabling swift and accurate prediction of protein structure from sequence. However, challenges persist in capturing sidechain packing,...
www.biorxiv.org
Alisia Fadini
@alisiafadini.bsky.social
· Feb 24
Alisia Fadini
@alisiafadini.bsky.social
· Feb 24