
https://mitchell-lab.umassmed.edu











doi.org/10.1038/s443...

doi.org/10.1038/s443...


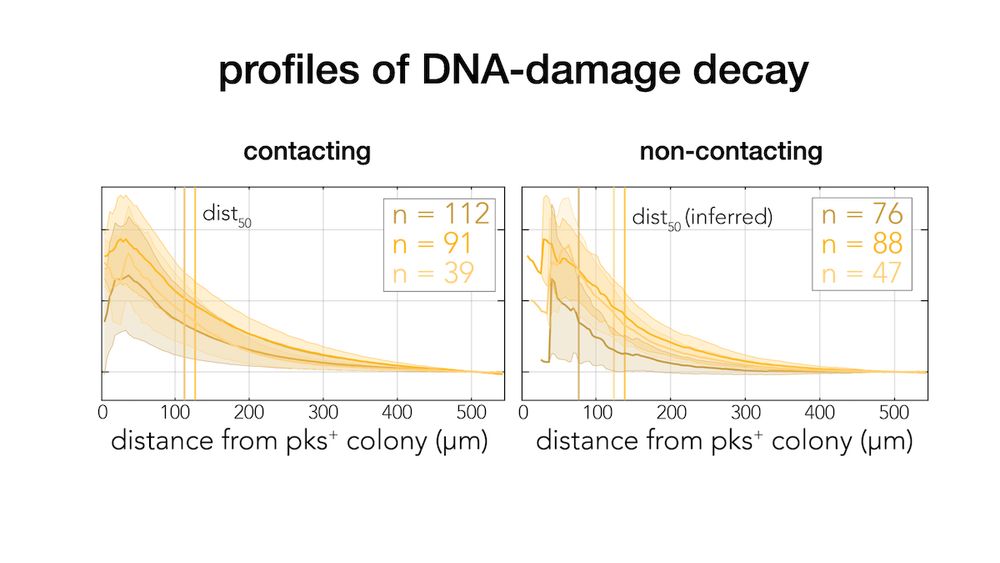
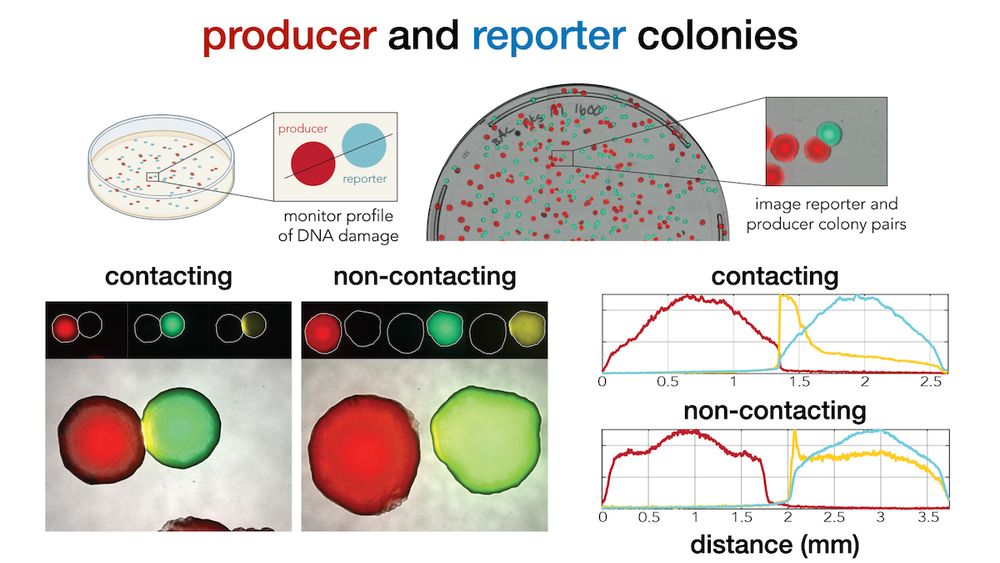
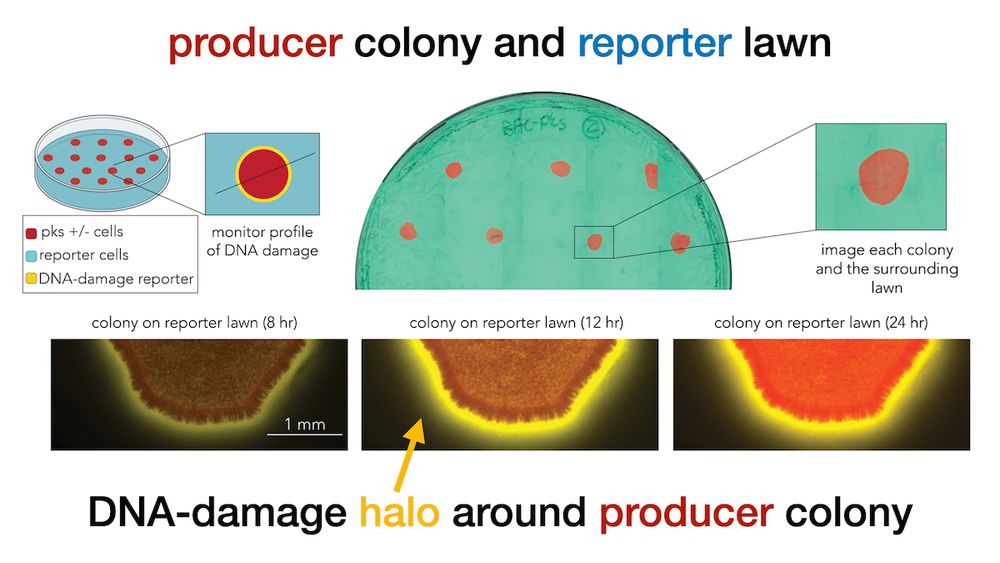
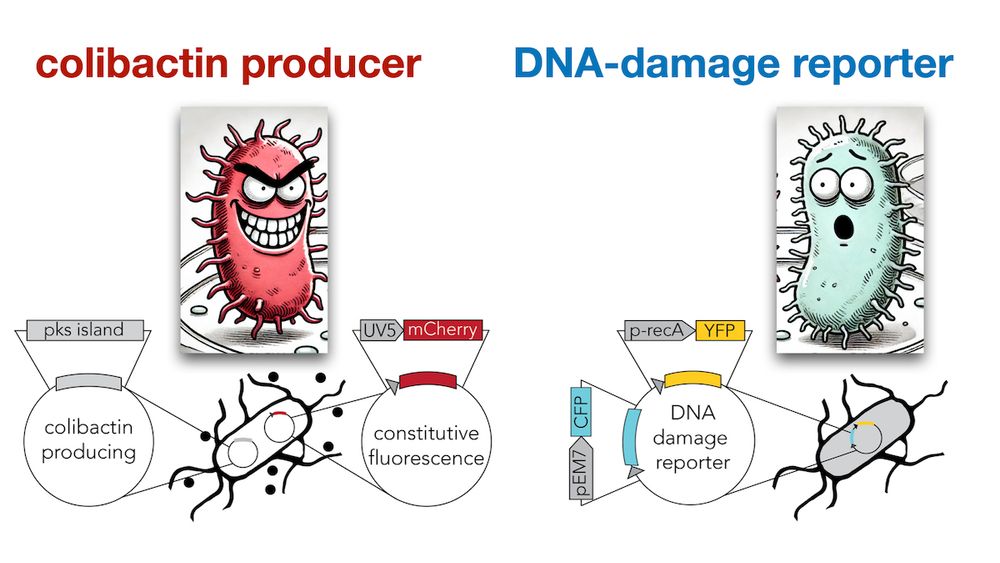
journals.asm.org/doi/10.1128/...
journals.asm.org/doi/10.1128/...






