Andreas Tolias
@andreastolias.bsky.social
730 followers
160 following
27 posts
Stanford Professor | NeuroAI Scientist | Entrepreneur working at the intersection of neuroscience, AI, and neurotechnology to decode intelligence @ enigmaproject.ai
Posts
Media
Videos
Starter Packs
Reposted by Andreas Tolias
Alexander Ecker
@aecker.bsky.social
· Apr 18

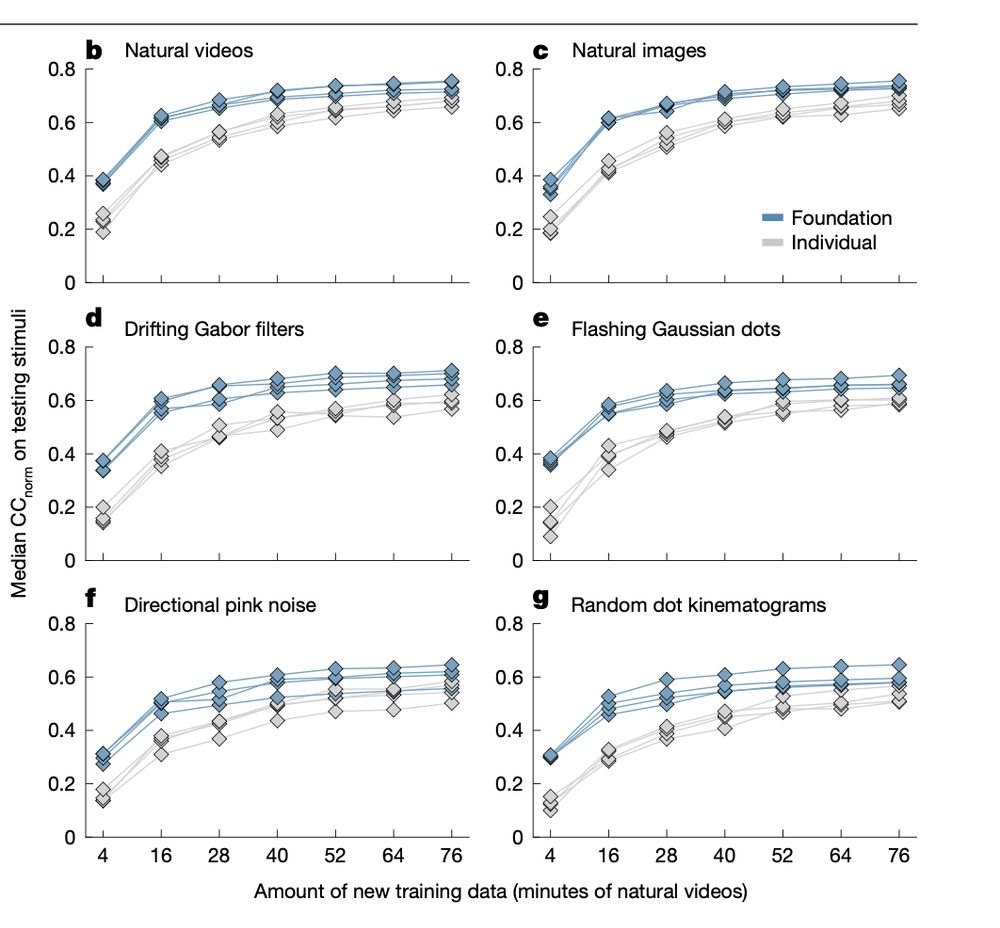
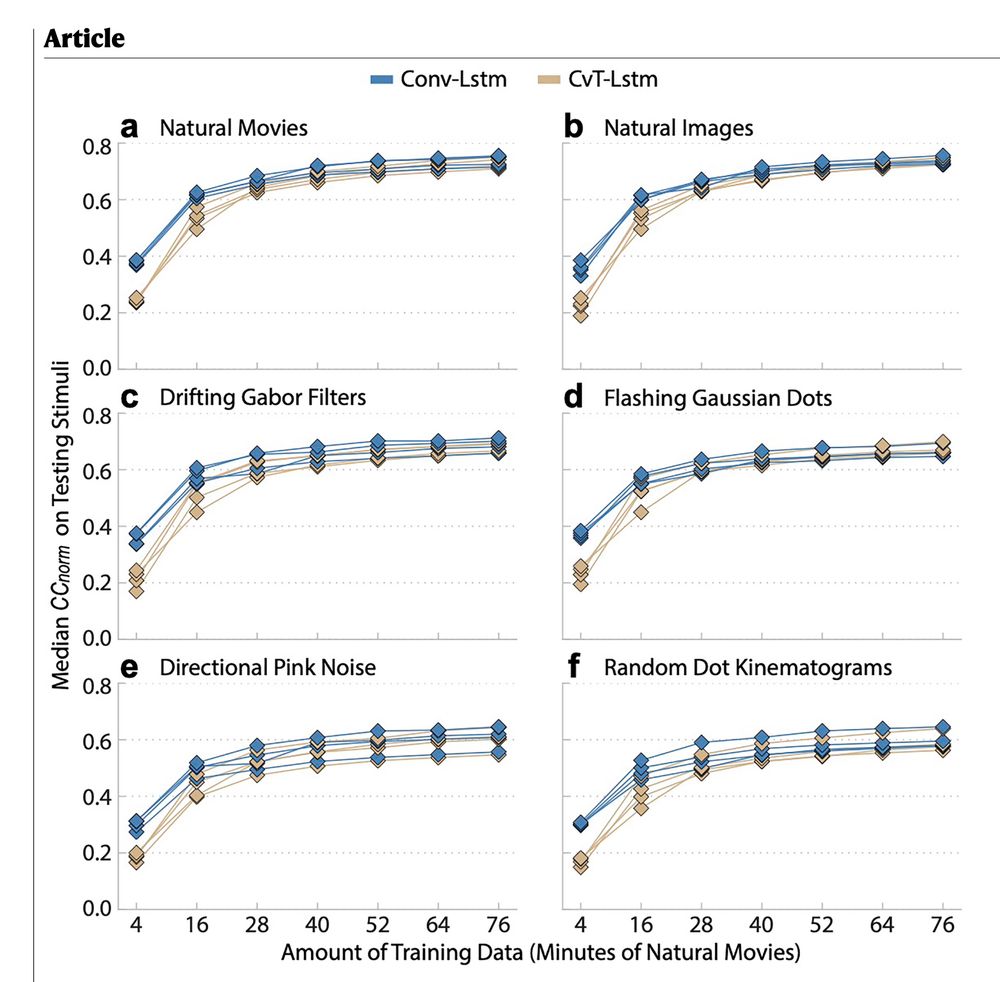
Generalization in data-driven models of primary visual cortex
Deep neural networks (DNN) have set new standards at predicting responses of neural populations to visual input. Most such DNNs consist of a convolutional network (core) shared across all neurons...
openreview.net
Andreas Tolias
@andreastolias.bsky.social
· Apr 15
Reposted by Andreas Tolias
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 13
Andreas Tolias
@andreastolias.bsky.social
· Apr 10
Andreas Tolias
@andreastolias.bsky.social
· Apr 10
Andreas Tolias
@andreastolias.bsky.social
· Apr 10