Anita Donlic
@anitadonlic.bsky.social
57 followers
27 following
24 posts
Postdoc in Brangwynne lab at Princeton, alumna of Hargrove lab. Working at the intersection of RNA chemical biology and condensates. Originally from Bosnia & Herzegovina
Posts
Media
Videos
Starter Packs
Anita Donlic
@anitadonlic.bsky.social
· Aug 20
Anita Donlic
@anitadonlic.bsky.social
· Aug 20
Anita Donlic
@anitadonlic.bsky.social
· Aug 20
Anita Donlic
@anitadonlic.bsky.social
· Aug 20

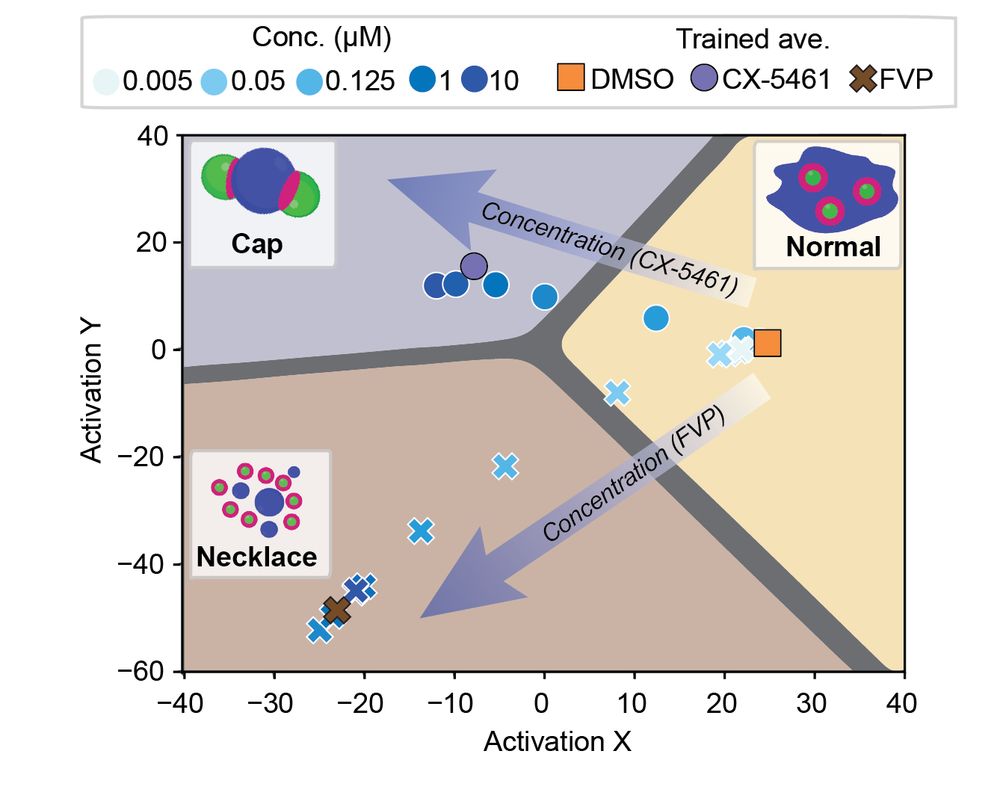
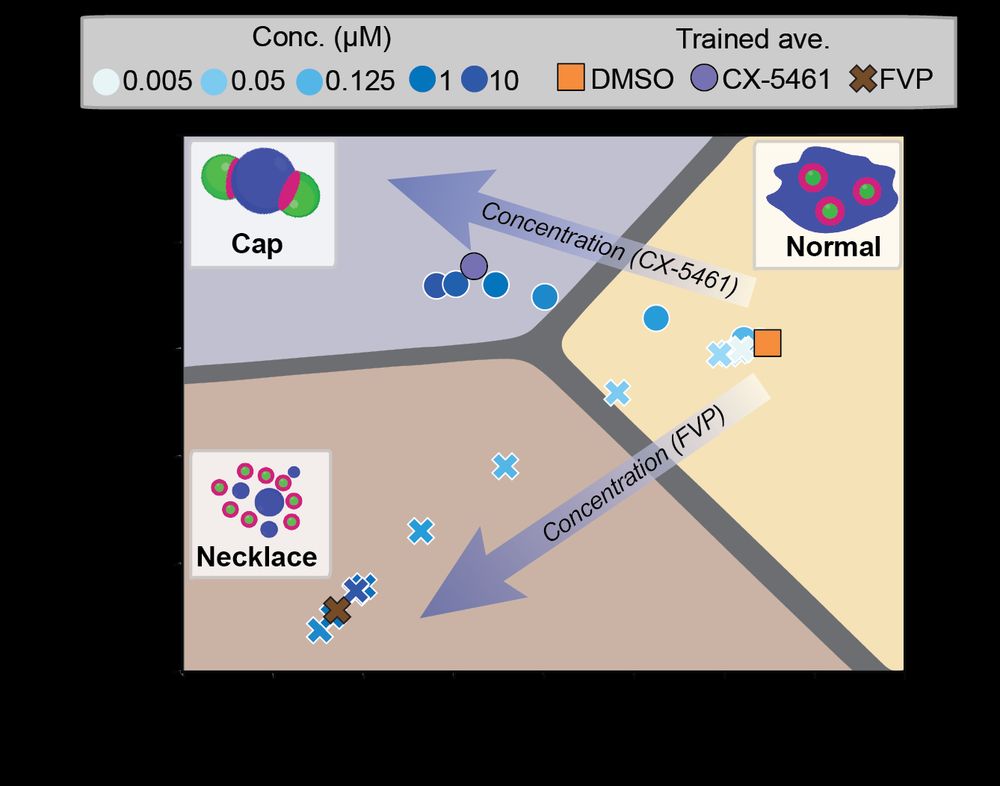
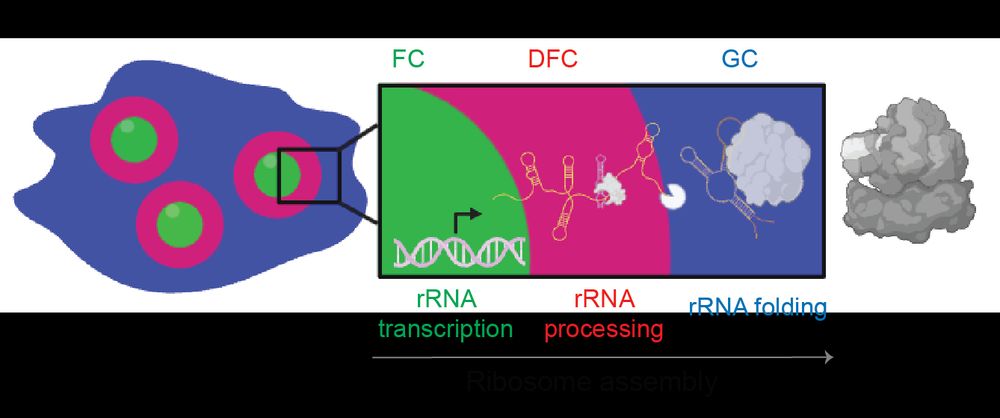
Deep Learning of Functional Perturbations from Condensate Morphology
Biomolecular condensates compartmentalize the interior of living cells to spatiotemporally organize complex functions, yet linking molecular interactions within condensates to their mesoscale organiza...
biorxiv.org
Anita Donlic
@anitadonlic.bsky.social
· Aug 20
Anita Donlic
@anitadonlic.bsky.social
· Aug 20
Anita Donlic
@anitadonlic.bsky.social
· Aug 20