Stephen Graham's Lab
@atomicvirology.bsky.social
2.2K followers
760 following
100 posts
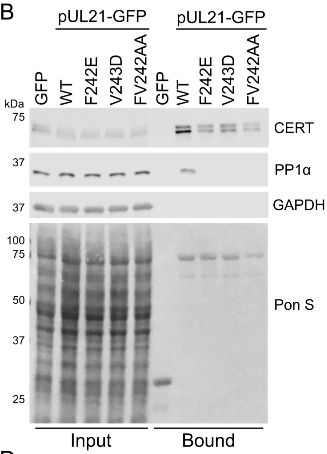
Lab in the University of Cambridge 🇬🇧 studying the cell biology, biophysics and structural biology of virus infection. Blame Stephen 🇦🇺 for the content.
Posts
Media
Videos
Starter Packs
Reposted by Stephen Graham's Lab
Chris H. Hill
@chillzaa.bsky.social
· Jul 19

A new protein-dependent riboswitch activates ribosomal frameshifting
Programmed -1 ribosomal frameshifting (PRF) is a ubiquitous translational control mechanism in RNA viruses, allowing them to change the relative abundance of proteins encoded in different reading fram...
www.biorxiv.org
Reposted by Stephen Graham's Lab
Chris H. Hill
@chillzaa.bsky.social
· Jul 19

A new protein-dependent riboswitch activates ribosomal frameshifting
Programmed -1 ribosomal frameshifting (PRF) is a ubiquitous translational control mechanism in RNA viruses, allowing them to change the relative abundance of proteins encoded in different reading fram...
www.biorxiv.org