Austin E. Y. T. Lefebvre
@austin-lefebvre.bsky.social
210 followers
200 following
26 posts
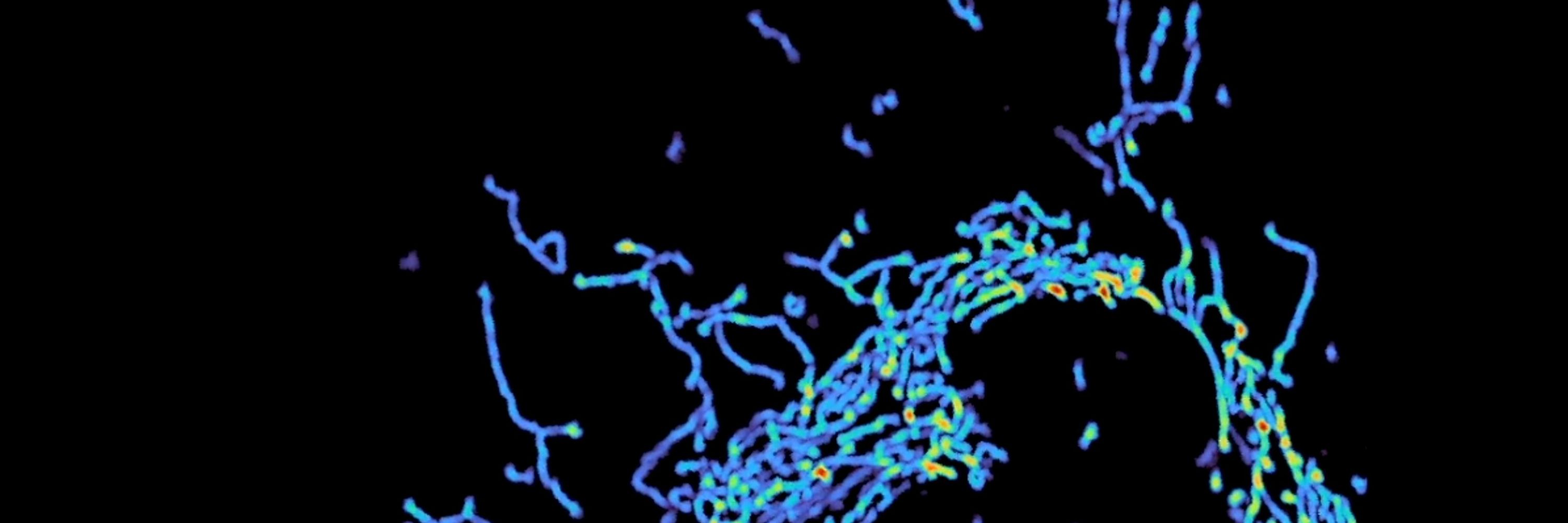
Dr. Leafbeaver, DS at Calico.
You can't spell imaging without "im aging" and boy am I.
Organelle enthusiast.
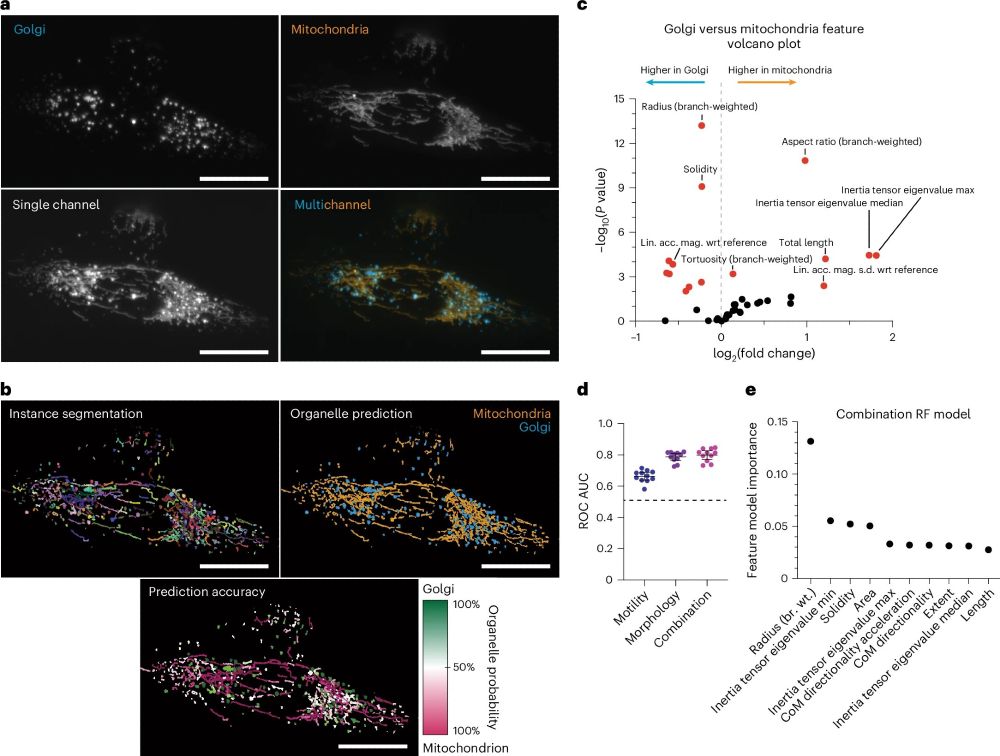
Creator of Nellie and Mitometer.
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Austin E. Y. T. Lefebvre
Reposted by Austin E. Y. T. Lefebvre
Rita Strack
@ritastrack.bsky.social
· Feb 28