Congratulations @jack-humphrey.bsky.social
www.nature.com/articles/s41...
(Collaborative work with Panos Roussos and his team! )
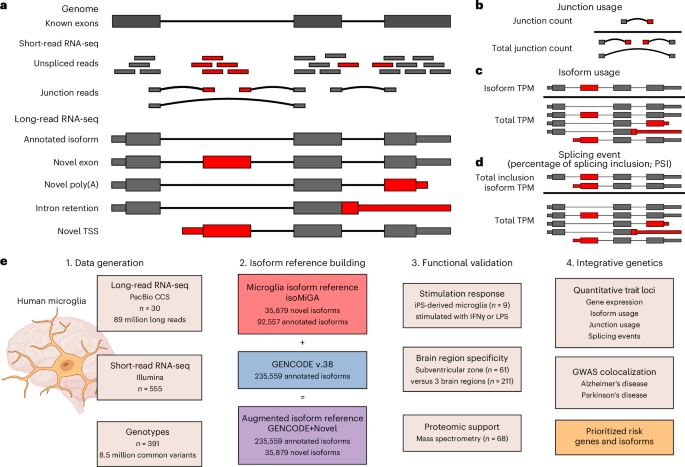
Long-read RNA sequencing atlas of human microglia isoforms elucidates disease-associated genetic regulation of splicing
Congratulations @jack-humphrey.bsky.social
www.nature.com/articles/s41...
(Collaborative work with Panos Roussos and his team! )
Long-read RNA sequencing atlas of human microglia isoforms elucidates disease-associated genetic regulation of splicing
doi.org/10.1101/2025.01.29.635391

doi.org/10.1101/2025.01.29.635391
www.biorxiv.org/content/10.1...

www.biorxiv.org/content/10.1...
Paper: www.nature.com/articles/s41...
Paper: www.nature.com/articles/s41...

