- #PD1 or #PDL1 expression didn't predict survival
- PD1 activity (from RIDDEN) did associate with survival.
Showing that receptor activity could be a more effective biomarker than expression. 5/n

- #PD1 or #PDL1 expression didn't predict survival
- PD1 activity (from RIDDEN) did associate with survival.
Showing that receptor activity could be a more effective biomarker than expression. 5/n



PLOS Computational Biology journals.plos.org/ploscompbiol... 1/n

PLOS Computational Biology journals.plos.org/ploscompbiol... 1/n

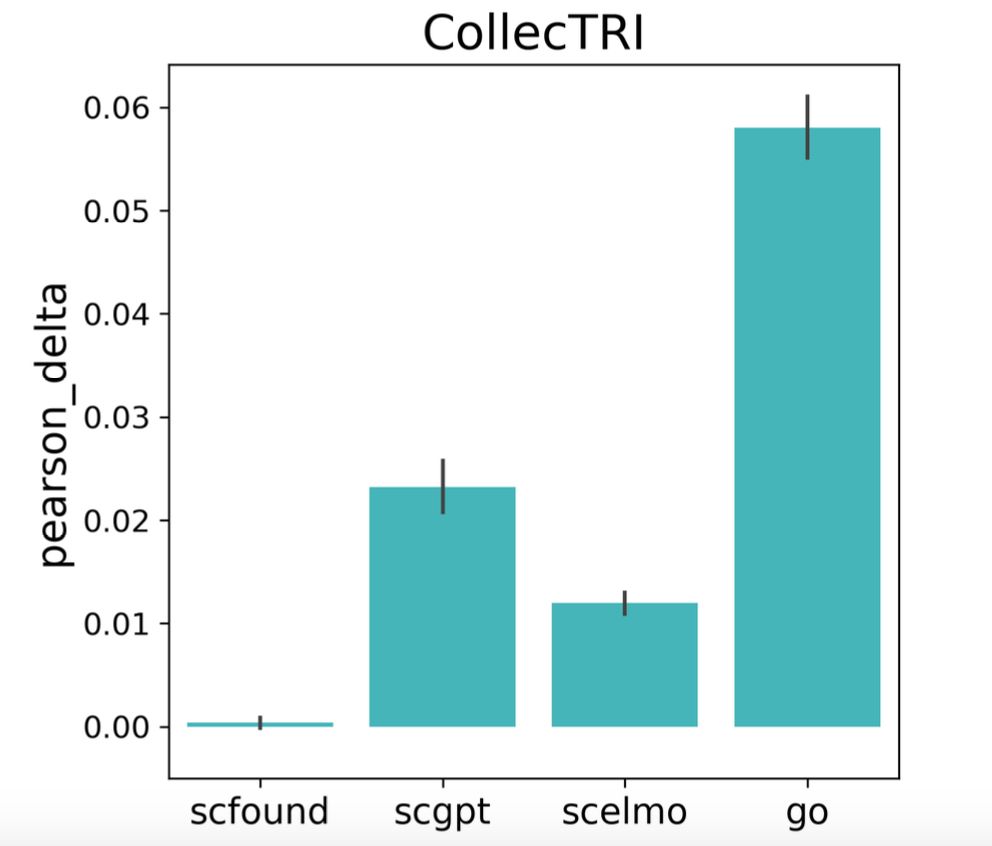

But how do they actually perform?
Our new paper from @turbine-ai.bsky.social is now out in BMC Genomics. bmcgenomics.biomedcentral.com/articles/10....
1/5

But how do they actually perform?
Our new paper from @turbine-ai.bsky.social is now out in BMC Genomics. bmcgenomics.biomedcentral.com/articles/10....
1/5
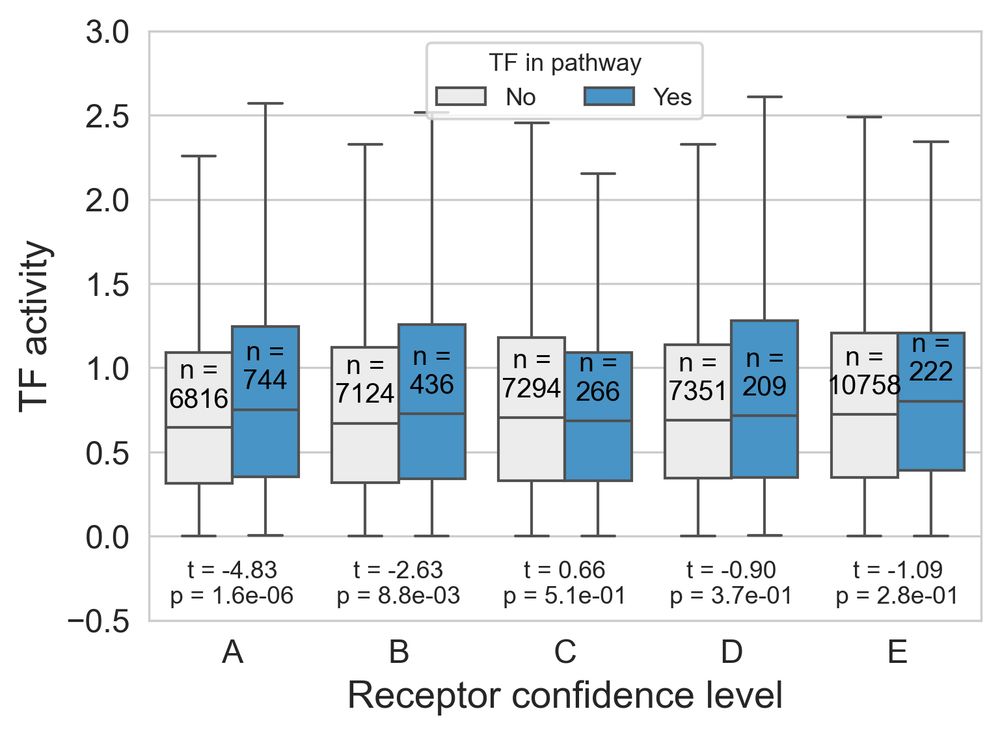
- PD1 or PDL1 expression didn't predict survival
- PD1 activity (from RIDDEN) did associate with survival.
This shows that receptor activity could be a more effective biomarker than expression. 5/n
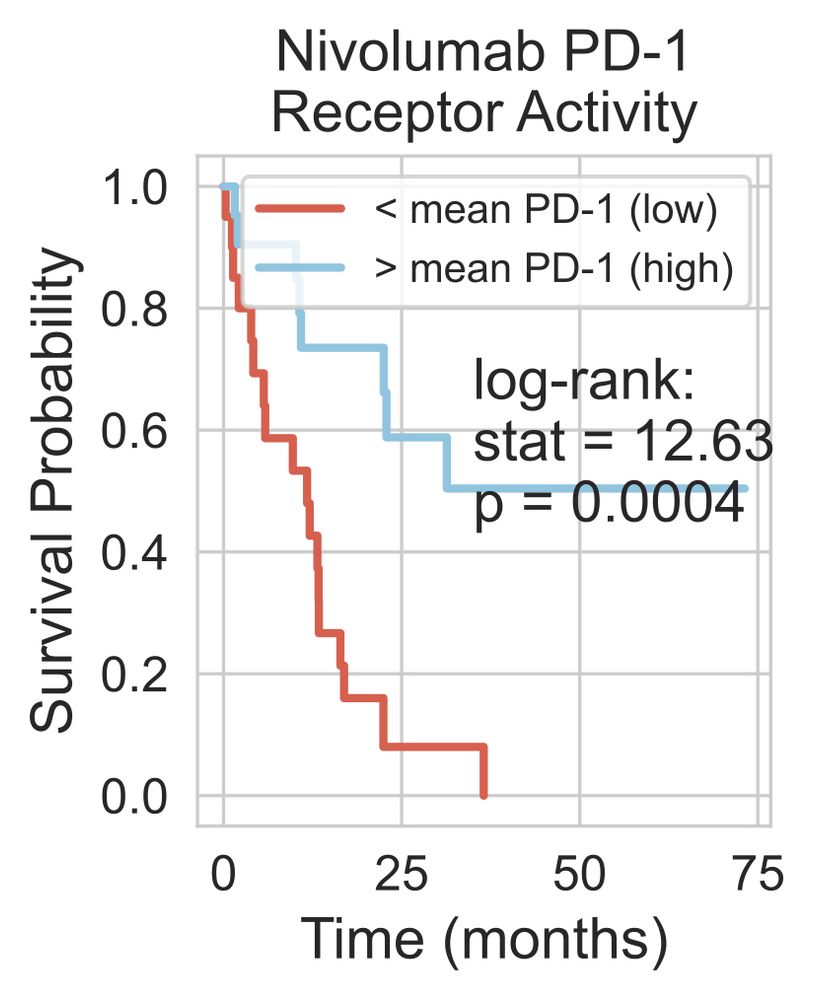
- PD1 or PDL1 expression didn't predict survival
- PD1 activity (from RIDDEN) did associate with survival.
This shows that receptor activity could be a more effective biomarker than expression. 5/n