BiaPy
@biapyx.bsky.social
320 followers
110 following
36 posts
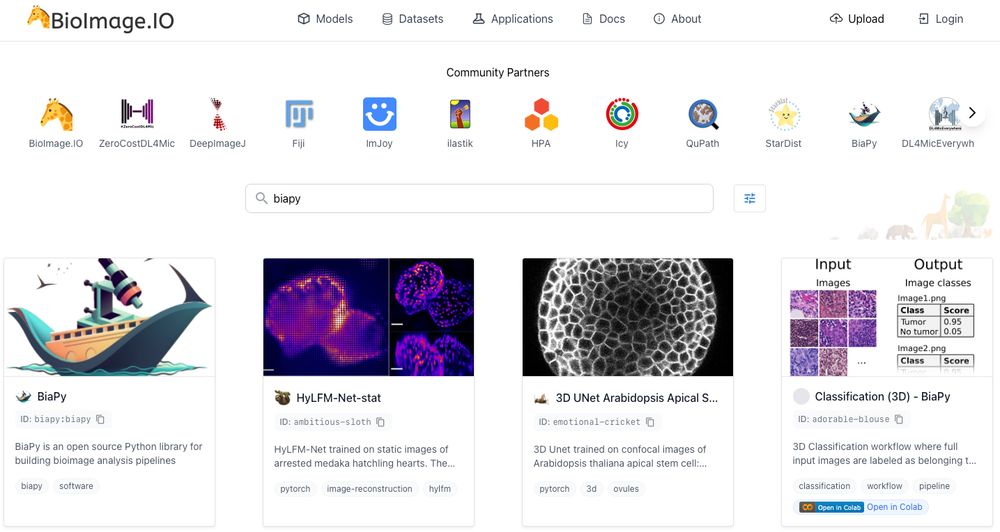
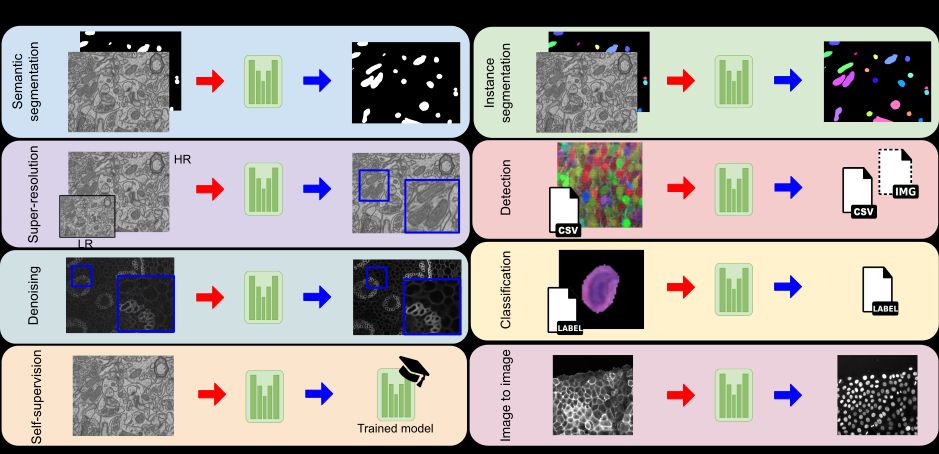
Accessible deep learning on bioimages
Site: https://biapyx.github.io/
Doc: https://biapy.readthedocs.io/en/latest/
Code: https://github.com/BiaPyX/BiaPy
Posts
Media
Videos
Starter Packs
Reposted by BiaPy
BiaPy
@biapyx.bsky.social
· May 2
BiaPy
@biapyx.bsky.social
· Apr 30
BiaPy
@biapyx.bsky.social
· Apr 30
BiaPy
@biapyx.bsky.social
· Apr 30