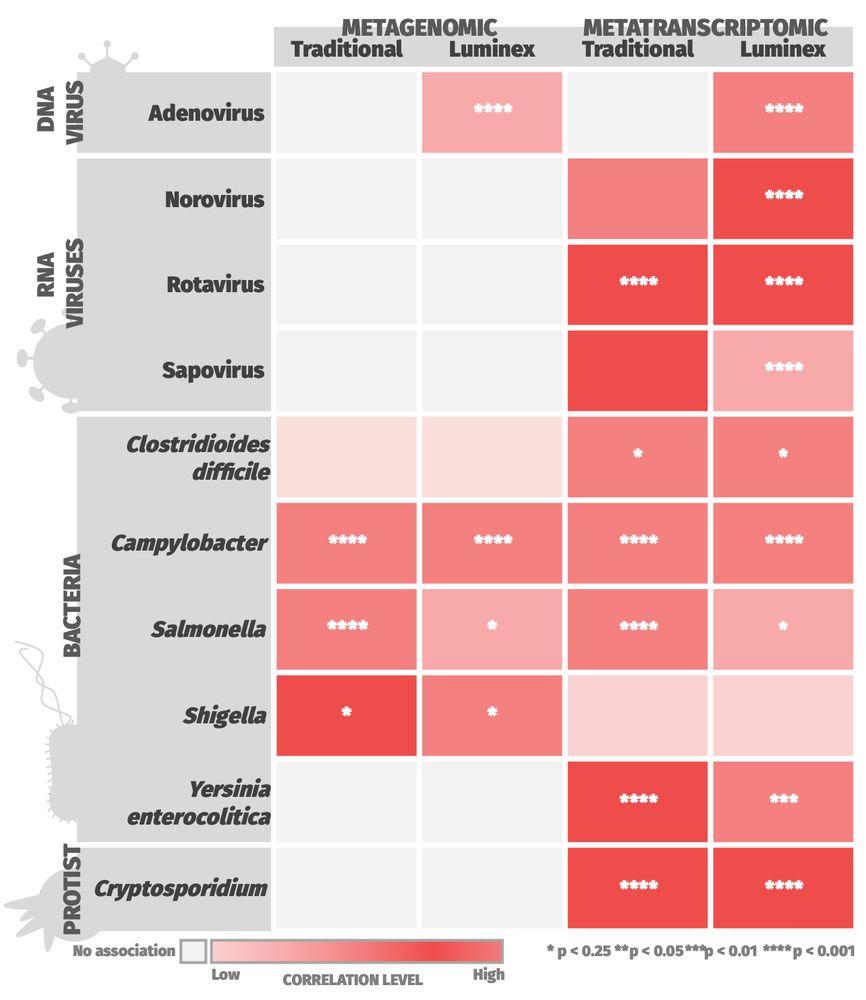
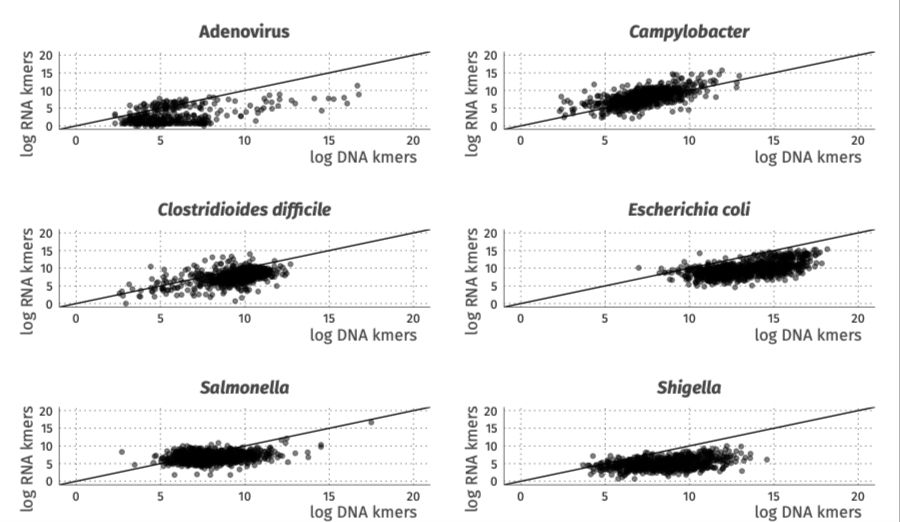
Dr Blanca Perez-Sepulveda
@blancaps.bsky.social
110 followers
120 following
4 posts
A tiny microbiologist working on big projects | iNTS | 10KSG | Phages | S. Panama | Hinton Lab | Chile | She/Her/Ella 🏳️🌈 views my own
Posts
Media
Videos
Starter Packs
Reposted by Dr Blanca Perez-Sepulveda
Kate Baker
@ksbakes.bsky.social
· Jul 27
Reposted by Dr Blanca Perez-Sepulveda
Kate Baker
@ksbakes.bsky.social
· Jul 27
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Ed Cunningham-Oakes
@edcoakes.bsky.social
· May 20
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda
Reposted by Dr Blanca Perez-Sepulveda