Posts
Media
Videos
Starter Packs
Reposted by Borna Novak
Alex Holehouse
@alexholehouse.bsky.social
· Feb 15
Reposted by Borna Novak
Borna Novak
@bornanovak.bsky.social
· Feb 15

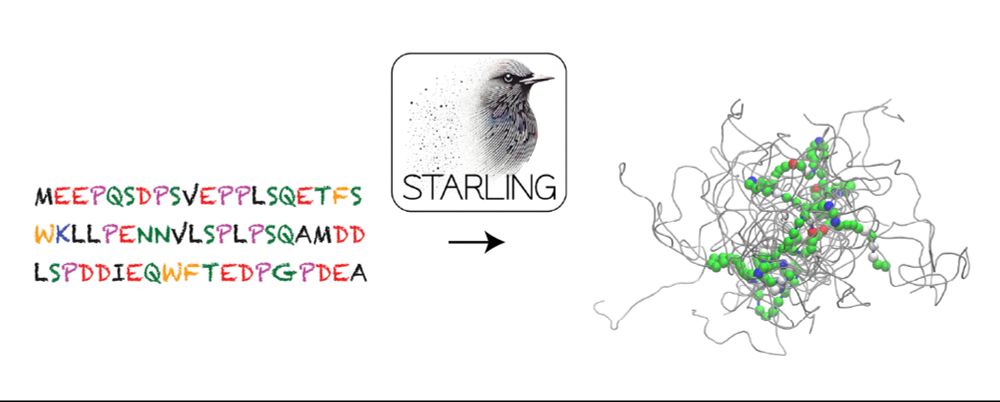
GitHub - idptools/starling: STARLING - conSTruction of intrinsicAlly disoRdered proteins ensembles efficientLy vIa multi-dimeNsional Generative models
STARLING - conSTruction of intrinsicAlly disoRdered proteins ensembles efficientLy vIa multi-dimeNsional Generative models - idptools/starling
github.com
Borna Novak
@bornanovak.bsky.social
· Feb 15
Borna Novak
@bornanovak.bsky.social
· Feb 15
Borna Novak
@bornanovak.bsky.social
· Feb 15
Borna Novak
@bornanovak.bsky.social
· Feb 15
Borna Novak
@bornanovak.bsky.social
· Feb 15
Borna Novak
@bornanovak.bsky.social
· Feb 15