Ben Valderrama
@bvalderrama.bsky.social
360 followers
350 following
36 posts
Computational Biologis at APC microbiome Ireland 🇮🇪
Personal website: https://rb.gy/455ftf
Microbiome-Gut-Brain axis | Gut permeability | Bioinformatics🦠🧬💻 | Stats
Posts
Media
Videos
Starter Packs
Pinned
Ben Valderrama
@bvalderrama.bsky.social
· Aug 11
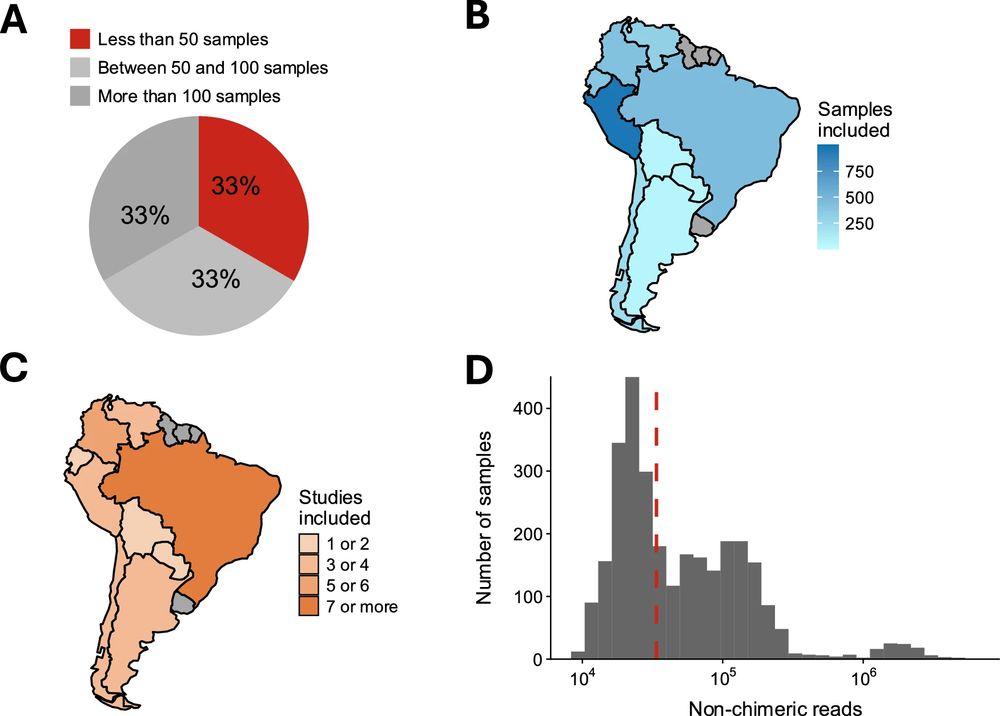
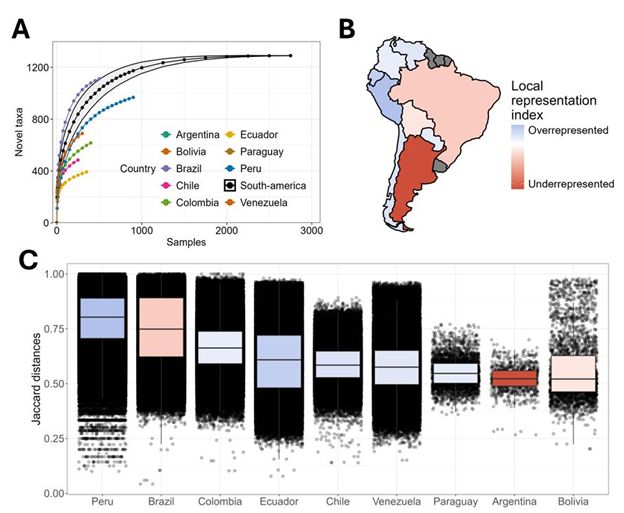
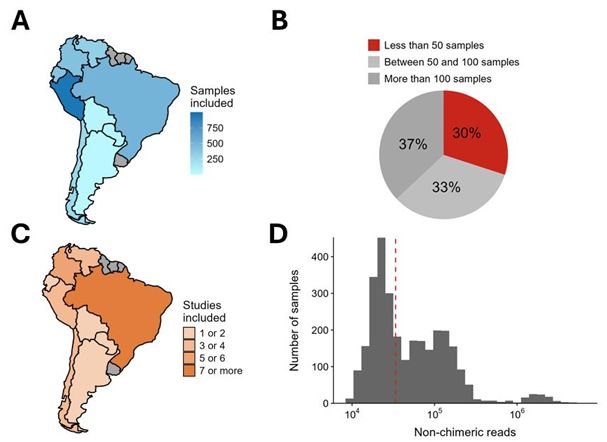
The South American MicroBiome Archive (saMBA): enriching the microbiome field by studying neglected populations - Nature Communications
Here, Valderrama et al., introduced ‘saMBA’, the largest collection of uniformly analyzed microbiome data from South America, the worlds most biodiverse yet less characterized region. The article prop...
shorturl.at
Reposted by Ben Valderrama
Reposted by Ben Valderrama
Ben Good
@benjaminhgood.bsky.social
· Aug 16

Prehistoric Global Migration of Vanishing Gut Microbes With Humans
The gut microbiome is crucial for health and greatly affected by lifestyle. Many microbes common in non-industrialized populations are disappearing or extinct in industrialized populations. Understand...
www.biorxiv.org
Reposted by Ben Valderrama
Ben Valderrama
@bvalderrama.bsky.social
· Aug 13
Reposted by Ben Valderrama
Ben Valderrama
@bvalderrama.bsky.social
· Aug 11
Reposted by Ben Valderrama
Ben Valderrama
@bvalderrama.bsky.social
· Aug 11
Ben Valderrama
@bvalderrama.bsky.social
· Aug 11

The South American MicroBiome Archive (saMBA): enriching the microbiome field by studying neglected populations - Nature Communications
Here, Valderrama et al., introduced ‘saMBA’, the largest collection of uniformly analyzed microbiome data from South America, the worlds most biodiverse yet less characterized region. The article prop...
shorturl.at
Reposted by Ben Valderrama
María R. Aburto
@maburto.bsky.social
· Aug 6

Gut microbiota regulates exercise-induced hormetic modulation of cognitive function
These findings suggest that the hormetic effects of physical exercise on cognitive
function and neurogenesis are mediated by corresponding changes in the gut microbiota,
highlighting a novel mechanist...
www.thelancet.com
Ben Valderrama
@bvalderrama.bsky.social
· Jul 21

Common Limitations of Gut Microbiome Meta-Analyses Undermine their Credibility
Although microbiome meta-analyses are fairly common in the literature, there is a lack of critical discussion on their limitations. This post is my (unsolicited) contribution to that topic.
shorturl.at
Reposted by Ben Valderrama
Reposted by Ben Valderrama
Lior Pachter
@lpachter.bsky.social
· Jun 16
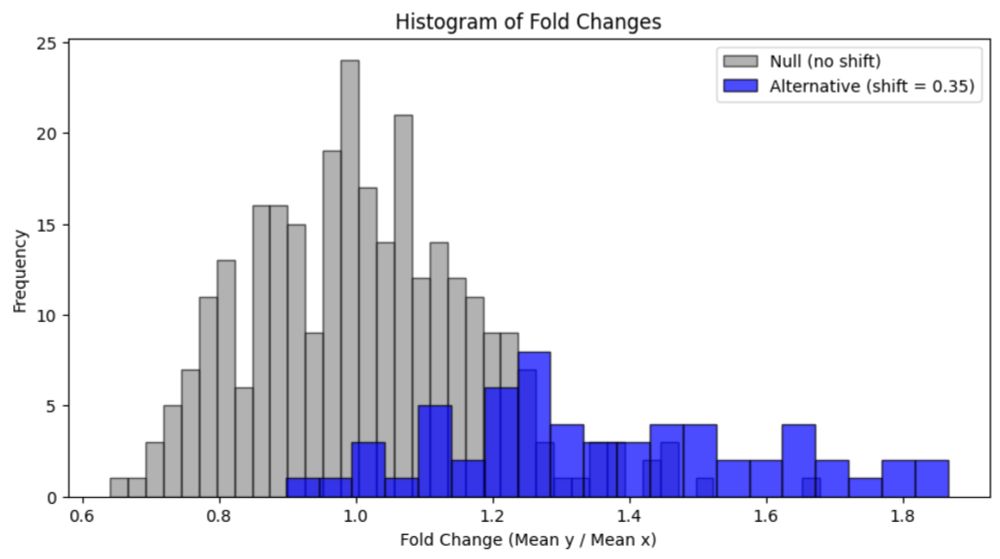
Reply to: Reply to: False positives in the study of memory-related gene expression
In the Nature paper “Spatial transcriptomics reveal neuron–astrocyte synergy in long-term memory” published on March 14th, 2024, authors Sun et al. claimed to identify cell-type specifi…
liorpachter.wordpress.com
Reposted by Ben Valderrama
Rich Abdill
@richabdill.com
· May 19

Compendium Manager: a tool for coordination of workflow management instances for bulk data processing in Python
Compendium Manager is a command-line tool written in Python to automate the provisioning, launch, and evaluation of bioinformatics pipelines. Although workflow management tools such as Snakemake and N...
arxiv.org
Reposted by Ben Valderrama
Mizrahi Lab
@mizrahilab.bsky.social
· Apr 21

The South American MicroBiome Archive (saMBA): Enriching the healthy microbiome concept by evaluating uniqueness and biodiversity of neglected populations
The composition and function of the human gut microbiome has been linked to multiple health outcomes across all world regions, often with region-specific associations. Unfortunately, the extent to whi...
www.biorxiv.org
Reposted by Ben Valderrama