Carli Peters
@carlipeters.bsky.social
950 followers
290 following
3 posts
Postdoctoral Researcher @ICArEHB interested in all things ancient proteins 🦴👩🏼🔬
Posts
Media
Videos
Starter Packs
Pinned
Carli Peters
@carlipeters.bsky.social
· Jun 3

Frontiers | Collagen peptide markers for three extinct Australian megafauna species
Recent advancements in biomolecular archaeology, such as stable isotope and ancient DNA research, have expanded our understanding of megafauna extinction pro...
www.frontiersin.org
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Sam Brown
@drsambrown.bsky.social
· Aug 28

MALDI Deamidation Score (MDS): A Fast and Flexible Method for Assessing Deamidation in ZooMS Data and Its Application to the Denisova Cave Bone Assemblage
Estimating deamidation from MALDI-TOF MS spectra of bones has most frequently been achieved using the q2e method due to its high-throughput capacity and ease of use. Despite this accessibility, q2e is...
www.biorxiv.org
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Anna Rufà
@annarb.bsky.social
· Jul 7
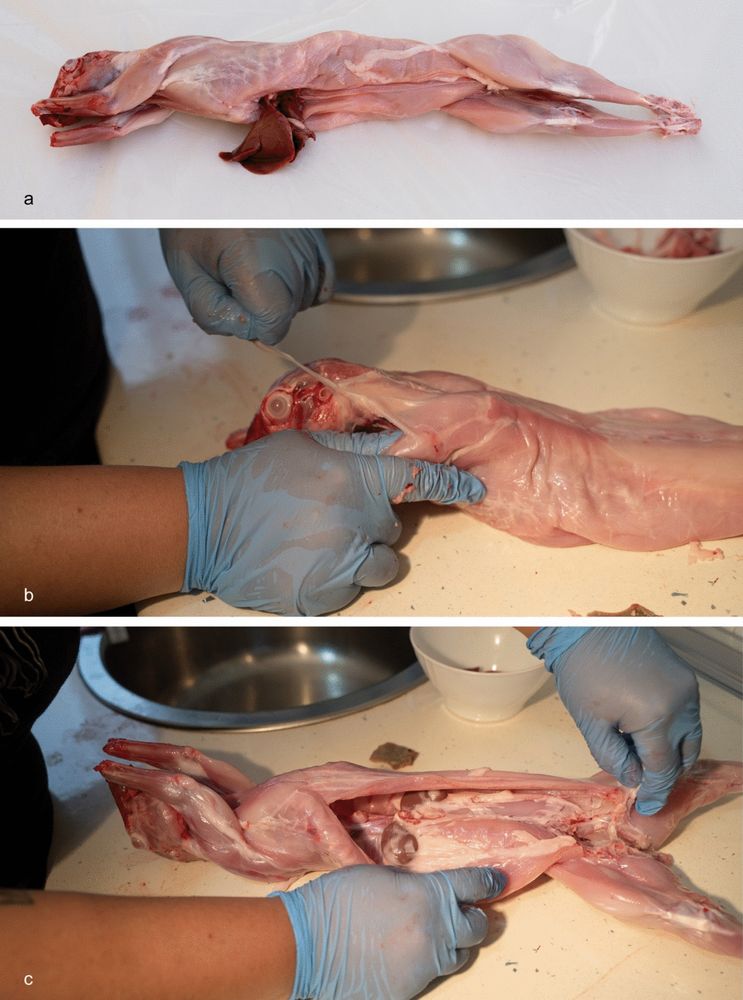
Experimental Protocol for Cooking Rabbits and its Archaeological Implications - Journal of Archaeological Method and Theory
Small prey such as rabbits are present in Middle Paleolithic and are abundant in the diet of Upper Paleolithic human groups in southwestern Europe, especially in the Iberian Peninsula. Several archaeo...
doi.org
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
Reposted by Carli Peters
ICArEHB
@icarehb.bsky.social
· Jun 5

Navigating the PhD journey amid prolonged conflict: Challenges, growth, and resilience
Author: Osman Khaleel // Editor: Cherene de Bruyn Cover image from Pexels *Archaeology: The study of past human cultures through the material culture (artefacts) left behind. *Lithics: Archaeologic…
research-hive.com