Carolin Klose
@carolinklose.bsky.social
110 followers
160 following
22 posts
PhD student in Munich
with Matthias Feige (TUM) and Brenda Schulman (MPI Biochemistry)
Proteostasis and membrane protein enthusiast
Boehringer Ingelheim Fonds fellow
Posts
Media
Videos
Starter Packs
Carolin Klose
@carolinklose.bsky.social
· Aug 19
Carolin Klose
@carolinklose.bsky.social
· Aug 13
Carolin Klose
@carolinklose.bsky.social
· Aug 12
Carolin Klose
@carolinklose.bsky.social
· Aug 11
Carolin Klose
@carolinklose.bsky.social
· Aug 11

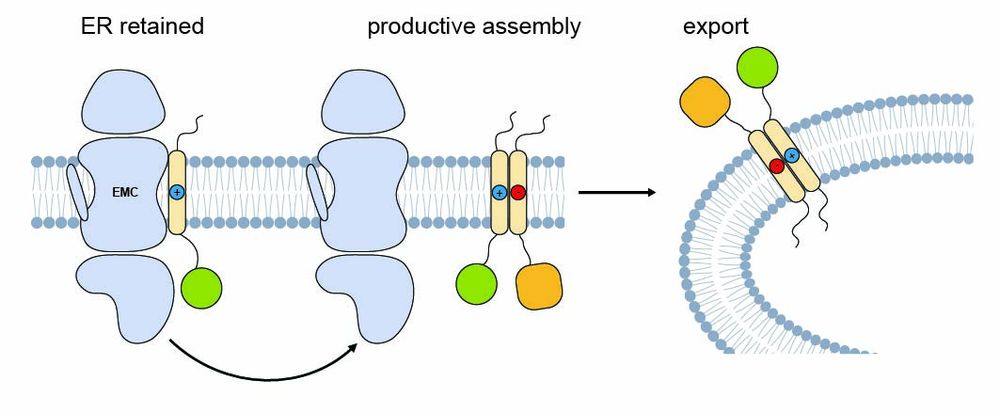
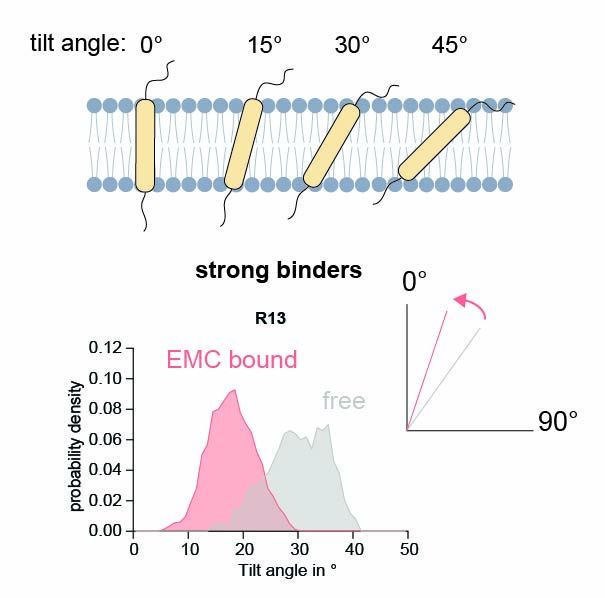
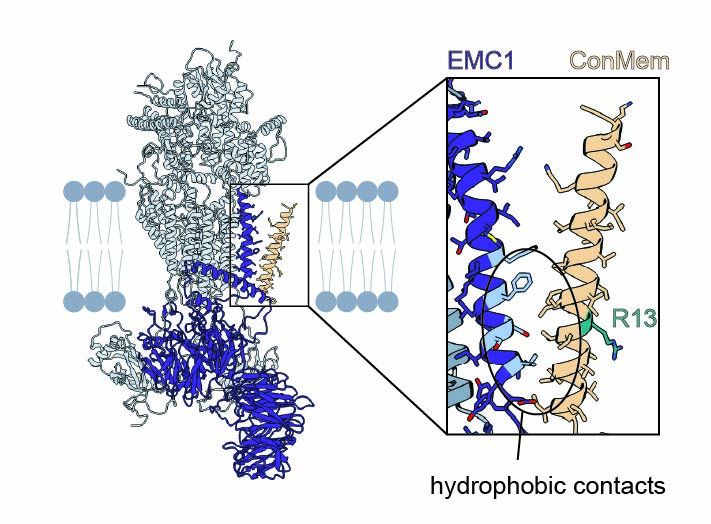
Structural basis of an EMC:Spf1 insertase-dislocase complex in the eukaryotic endoplasmic reticulum
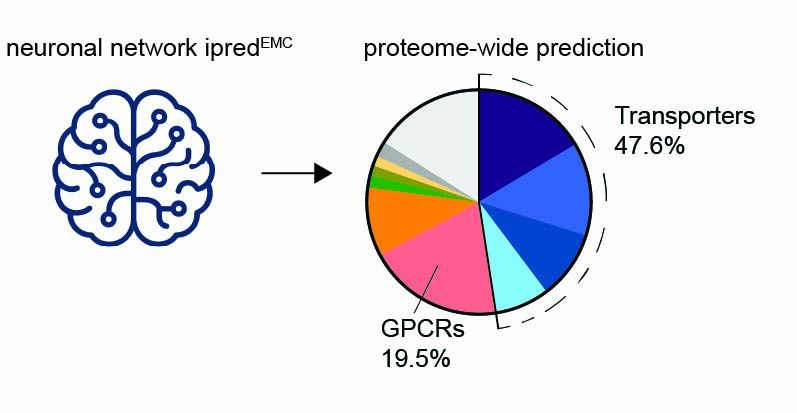
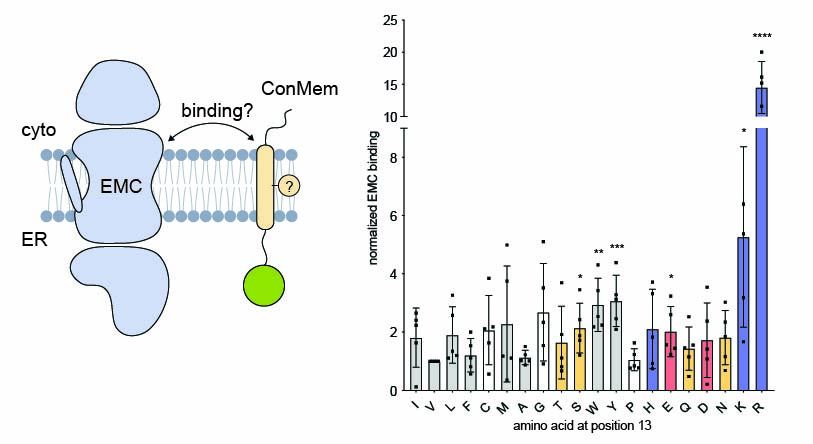
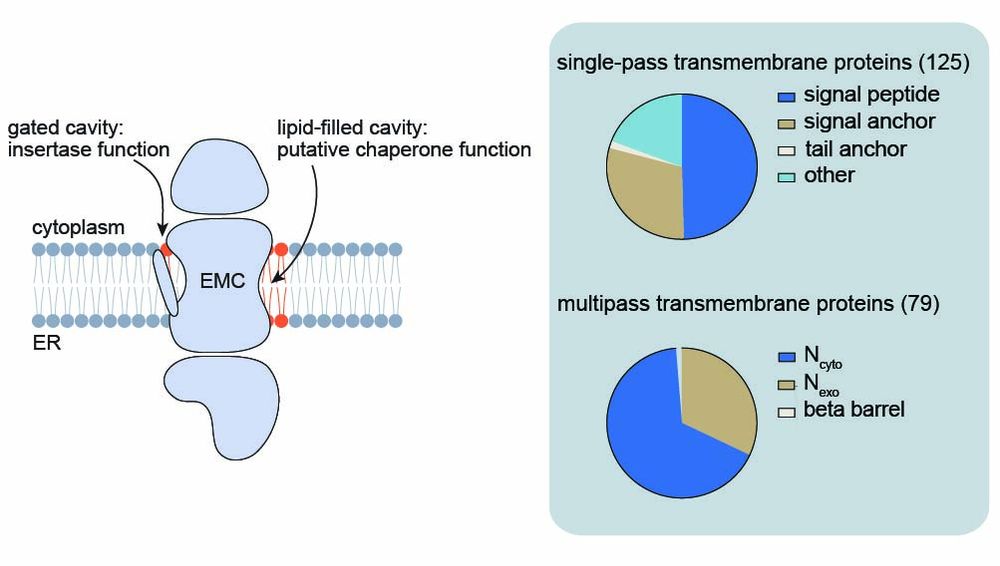
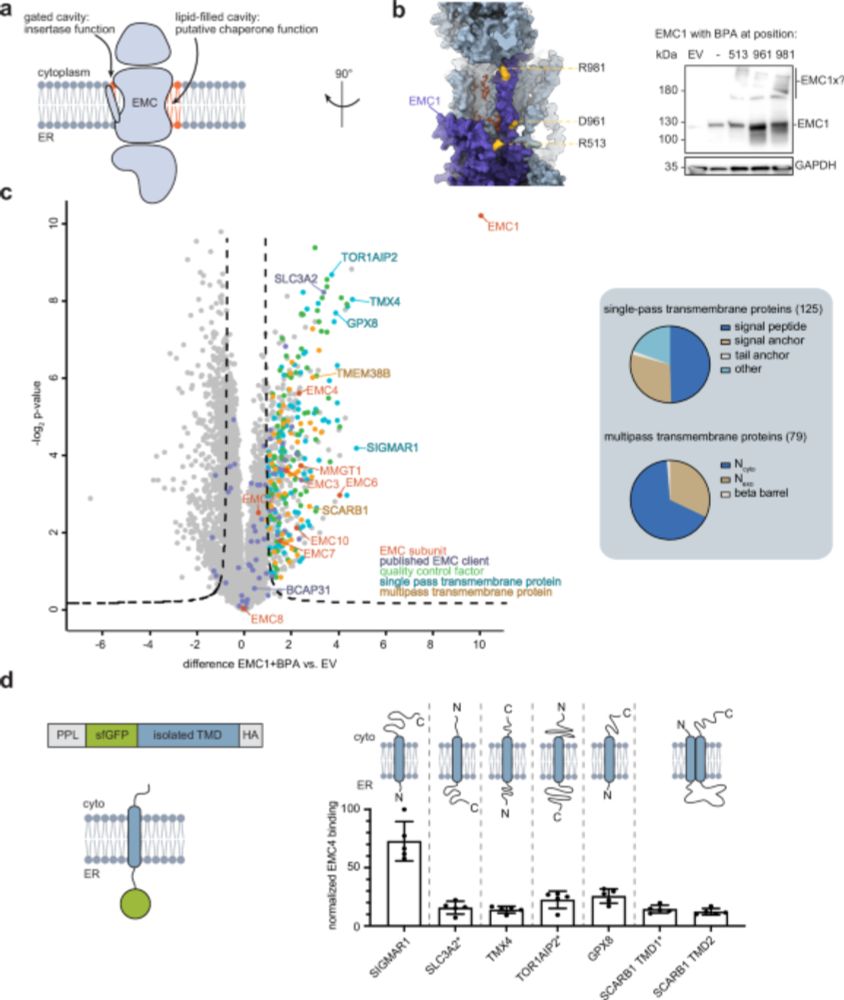
Most eukaryotic membrane proteins are inserted into the membrane at the endoplasmic reticulum (ER). This essential but error-prone process relies on molecular quality control machineries to prevent mi...
www.biorxiv.org
Carolin Klose
@carolinklose.bsky.social
· Mar 24