Excited to share recent work from our lab on noncoding genomics 🔎🧬: High-resolution CRISPR perturbations of the MYC TAD (~3 Mb) in 6 different human cancer cell lines
www.nature.com/articles/s41...

Excited to share recent work from our lab on noncoding genomics 🔎🧬: High-resolution CRISPR perturbations of the MYC TAD (~3 Mb) in 6 different human cancer cell lines
www.nature.com/articles/s41...
Here I want to expand on the value of using DIRECTIONAL information contained in LoF burden tests.🧵
[work led by @minetoota.bsky.social ]
bsky.app/profile/jkpr...
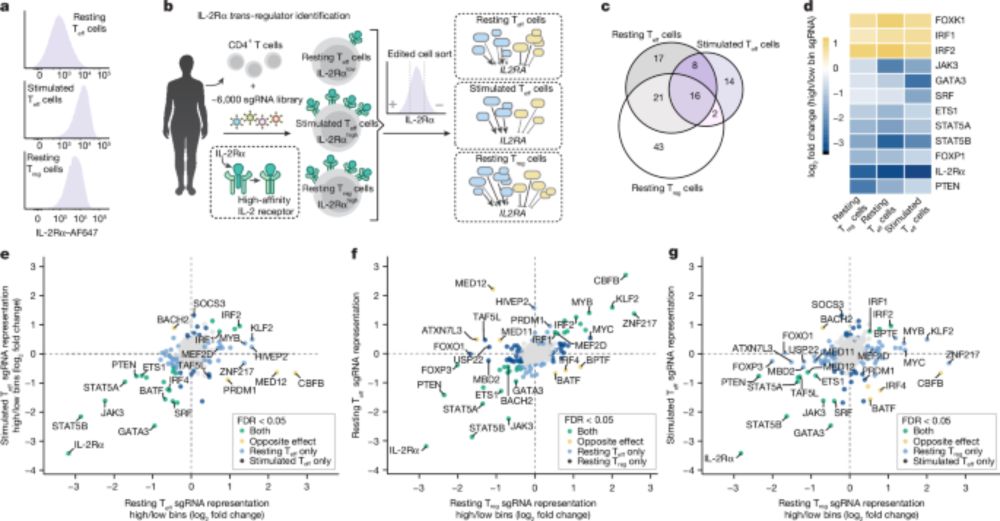
But interpretation is difficult as most effects flow through (unobserved) gene regulatory networks. Can we gain insight by linking to modern Perturb-seq data?
www.biorxiv.org/content/10.1...

Here I want to expand on the value of using DIRECTIONAL information contained in LoF burden tests.🧵
[work led by @minetoota.bsky.social ]
bsky.app/profile/jkpr...
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
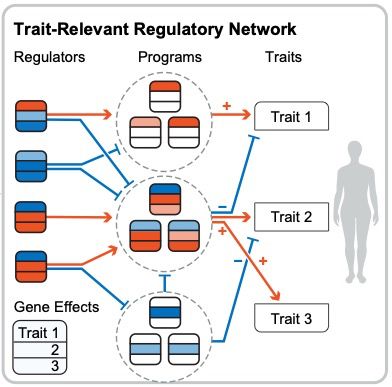
I'm very excited to present our new work combining associations and Perturb-seq to build interpretable causal graphs! A 🧵
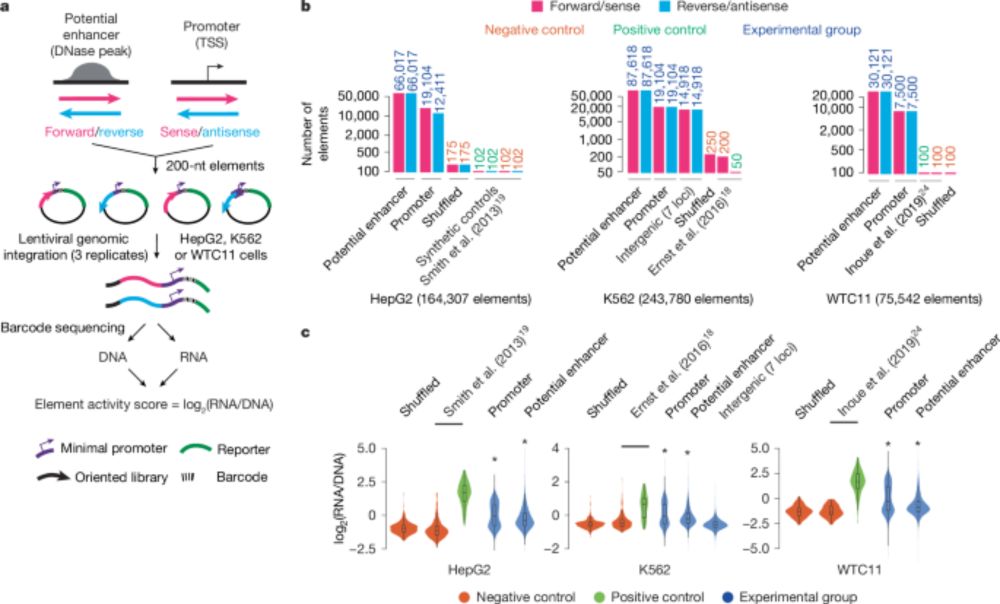
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
www.biorxiv.org/content/10.1...
Huge congrats to Anusri! This was quite a slog (for both of us) but we r very proud of this one! It is a long read but worth it IMHO. Methods r in the supp. materials. Bluetorial coming soon below 1/
Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...

Do these studies find the most IMPORTANT genes? If not, how DO they rank genes?
Here we present a surprising result: these studies actually test for SPECIFICITY! A 🧵on what this means... (🧪🧬)
www.biorxiv.org/content/10.1...
www.nature.com/articles/s41...
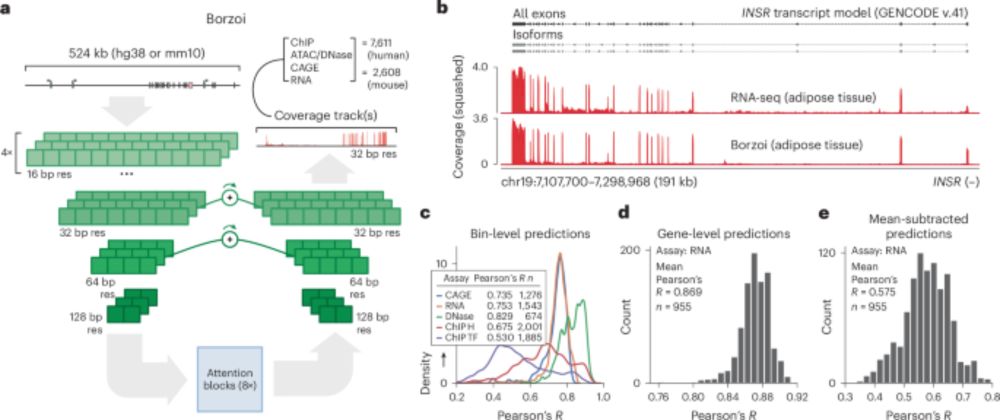
www.nature.com/articles/s41...

