Chris Rinke
@chrisrinke.bsky.social
410 followers
80 following
27 posts
Professor of Microbiology @ University of Innsbruck, Austria
Honorary Associate Professor @ The University of Queensland, Australia
Environmental OMICS (E-OMICS) team lead
http://www.rinkelab.org
(former twitter/x account: @ChristianRinke )
Posts
Media
Videos
Starter Packs
Chris Rinke
@chrisrinke.bsky.social
· Apr 27
Chris Rinke
@chrisrinke.bsky.social
· Apr 27
Chris Rinke
@chrisrinke.bsky.social
· Apr 27
Chris Rinke
@chrisrinke.bsky.social
· Apr 27

Metagenomic insights into taxonomic and functional patterns in shallow coastal and deep subseafloor sediments in the Western Pacific
Marine sediments are vast, underexplored habitats and represent one of the largest carbon deposits on our planet. Microbial communities drive nutrient cycling in these sediments, but the full extent o...
www.microbiologyresearch.org
Reposted by Chris Rinke
Chris Rinke
@chrisrinke.bsky.social
· Apr 3
Chris Rinke
@chrisrinke.bsky.social
· Apr 3
Chris Rinke
@chrisrinke.bsky.social
· Mar 13
Chris Rinke
@chrisrinke.bsky.social
· Mar 13
Chris Rinke
@chrisrinke.bsky.social
· Mar 13
Reposted by Chris Rinke
SeqCode
@seqcode.bsky.social
· Feb 11
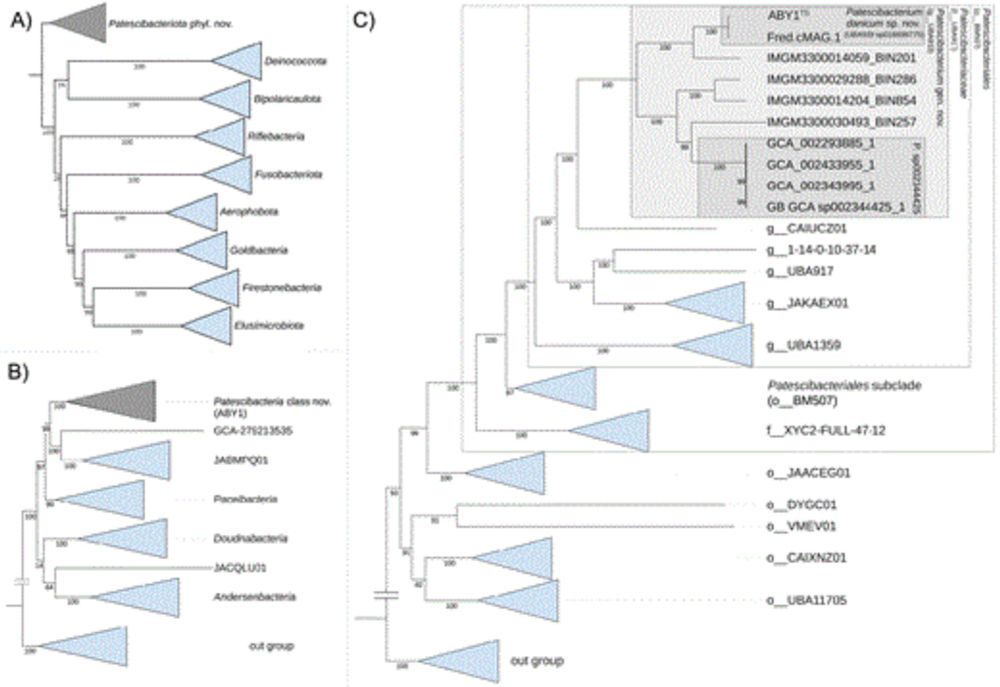
Proposal of Patescibacterium danicum gen. nov., sp. nov. in the ubiquitous bacterial phylum Patescibacteriota phyl. nov.
Abstract. Candidatus Patescibacteria is a diverse bacterial phylum that is notable for members with ultrasmall cell size, reduced genomes, limited metaboli
doi.org
Chris Rinke
@chrisrinke.bsky.social
· Feb 8
Chris Rinke
@chrisrinke.bsky.social
· Feb 8
Chris Rinke
@chrisrinke.bsky.social
· Feb 8

Proposal of Patescibacterium danicum gen. Nov., sp. nov. in the ubiquitous ultrasmall bacterial phylum Patescibacteriota phyl. Nov
Abstract. Candidatus Patescibacteria is a diverse bacterial phylum that is notable for members with ultrasmall cell size, reduced genomes, limited metaboli
doi.org
Chris Rinke
@chrisrinke.bsky.social
· Feb 8
Chris Rinke
@chrisrinke.bsky.social
· Jan 29
Chris Rinke
@chrisrinke.bsky.social
· Jan 8

Apoorva Prabhu (@aprabhu90.bsky.social)
Postdoctoral Research Associate - Sydney Institute of Marine Science
Honorary Postdoc- University of Technology, Sydney
PhD at the Australian Centre for Ecogenomics, The University of Queensland, Bri...
aprabhu90.bsky.social
Chris Rinke
@chrisrinke.bsky.social
· Jan 8

Insights Into Phylogeny, Diversity and Functional Potential of Poseidoniales Viruses
Our spatio-temporal metagenomic study of a subtropical estuary successfully identified viruses and their potential hosts. Viral contigs were detected using a consensus approach comprising geNOMAD and...
doi.org
Chris Rinke
@chrisrinke.bsky.social
· Nov 19
Chris Rinke
@chrisrinke.bsky.social
· Nov 18



