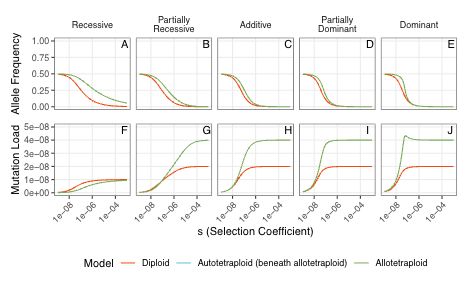
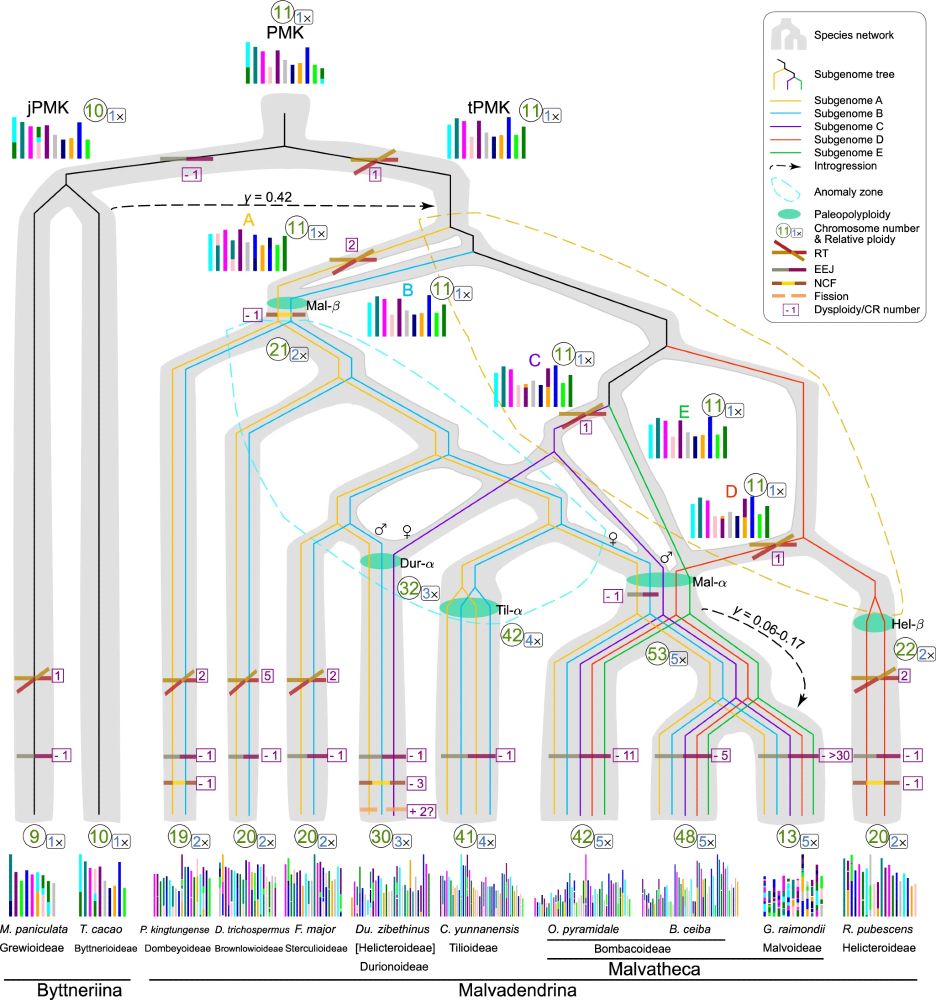
Justin Conover
@conjustover.bsky.social
1.1K followers
1.7K following
57 posts
Assistant Member and Principal Investigator
Donald Danforth Plant Science Center, St. Louis MO
Interested in plants, popgen, polyploidy, and puns.
Posts
Media
Videos
Starter Packs
Justin Conover
@conjustover.bsky.social
· Aug 21
Justin Conover
@conjustover.bsky.social
· Aug 20
Justin Conover
@conjustover.bsky.social
· Aug 20
Justin Conover
@conjustover.bsky.social
· Aug 13
Justin Conover
@conjustover.bsky.social
· Aug 13
Justin Conover
@conjustover.bsky.social
· Aug 13
Justin Conover
@conjustover.bsky.social
· Aug 13
Justin Conover
@conjustover.bsky.social
· Aug 13
Justin Conover
@conjustover.bsky.social
· Aug 13