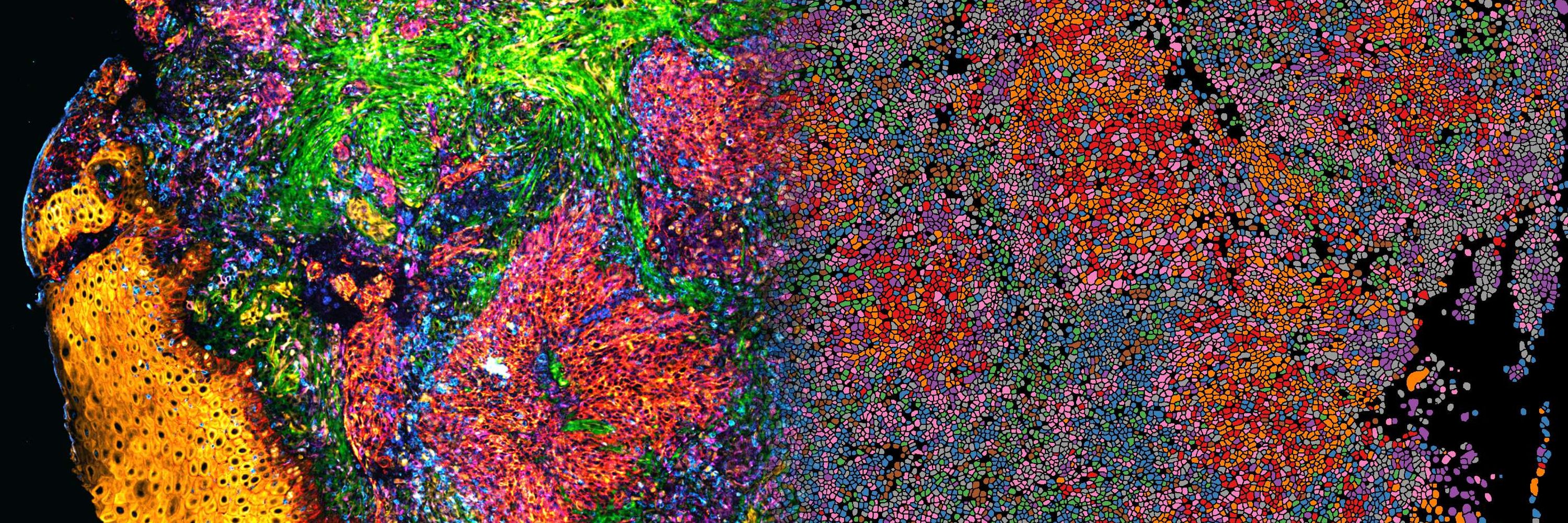Coscia Lab
@coscialab.bsky.social
210 followers
74 following
9 posts
...we are passionate about spatial proteomics, mass spectrometry and single-cell technologies to study disease mechanisms @MDC_Berlin 🇩🇪🇨🇴🇨🇳🇰🇿🇵🇷🇵🇱🇲🇽
Posts
Media
Videos
Starter Packs
Coscia Lab
@coscialab.bsky.social
· Jun 23
Coscia Lab
@coscialab.bsky.social
· Jun 5

An ultrasensitive spatial tissue proteomics workflow exceeding 100 proteomes per day
Achieving high-resolution spatial tissue proteomes requires careful balancing and integration of optimized sample processing, chromatography, and MS acquisition. Here, we present an advanced cellenONE...
bit.ly
Coscia Lab
@coscialab.bsky.social
· Mar 20

Deep spatial proteomics of ovarian cancer precursor lesions delineates early disease changes and cell-of-origin signatures
High-grade serous ovarian cancer (HGSOC) is a devastating disease that is frequently detected at an advanced and incurable stage. Advances in ultrasensitive mass spectrometry-based spatial proteomics ...
www.biorxiv.org
Reposted by Coscia Lab
Reposted by Coscia Lab
Reposted by Coscia Lab
Coscia Lab
@coscialab.bsky.social
· Jan 30
Coscia Lab
@coscialab.bsky.social
· Dec 8
Rita Strack
@ritastrack.bsky.social
· Dec 6

Method of the Year 2024: spatial proteomics - Nature Methods
Approaches for profiling the spatial proteome in tissues are the basis of atlas-scale projects that are delivering on their promise for understanding biological complexity in health and disease.
www.nature.com