David Gokhman
@david-gokhman.bsky.social
290 followers
240 following
31 posts
Asst Prof @WeizmannScience. Exploring human evolution through the lens of gene regulation🧬 Still rooting for Neanderthals & Denisovans💀❤️ gokhmanlab.com
Posts
Media
Videos
Starter Packs
Pinned
David Gokhman
@david-gokhman.bsky.social
· Aug 28
David Gokhman
@david-gokhman.bsky.social
· Aug 27
David Gokhman
@david-gokhman.bsky.social
· Aug 27

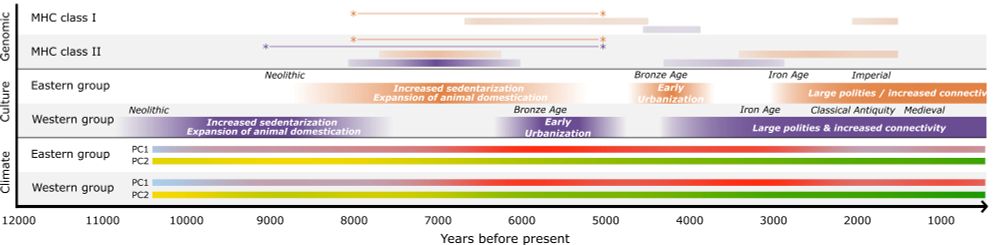
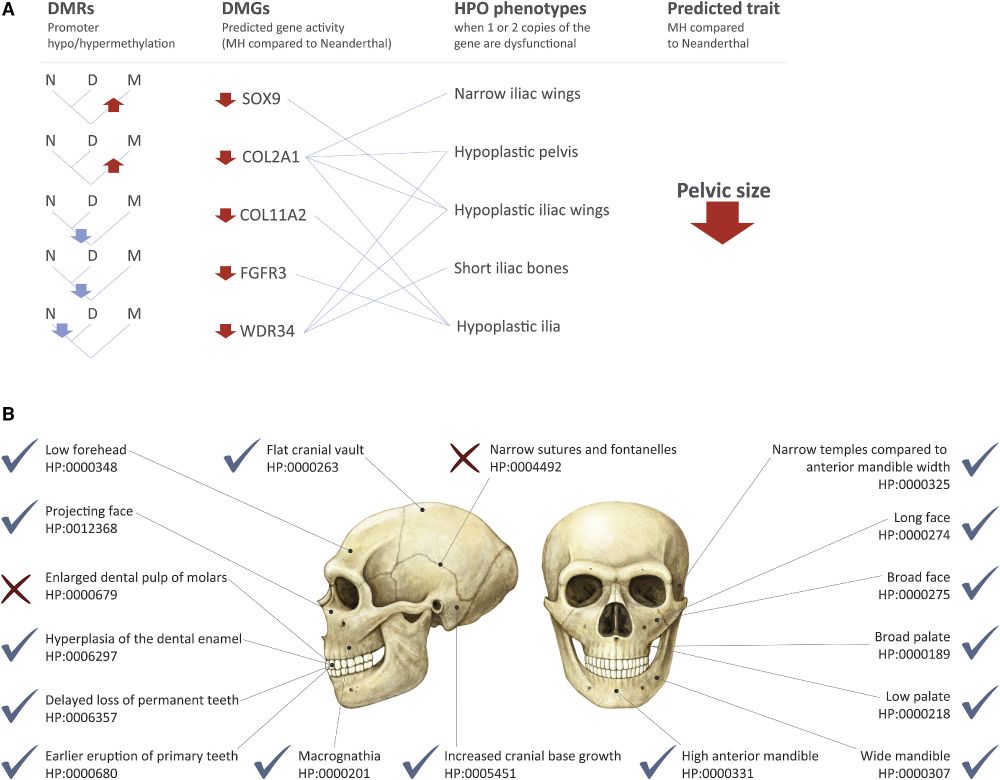
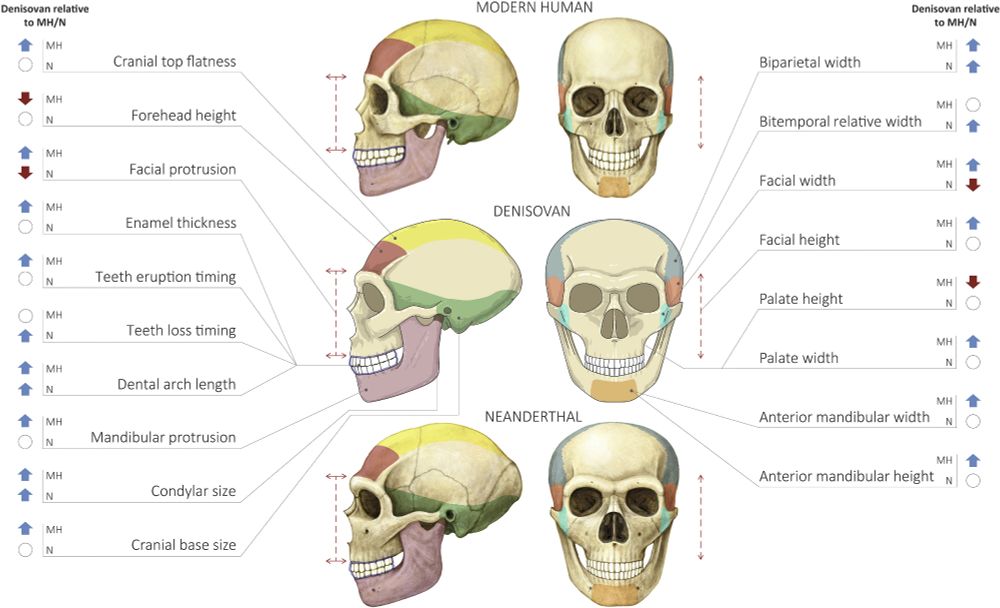
Predicting the direction of phenotypic difference - Nature Communications
Here authors reveal a method to predict key information on phenotypes - their direction. This is achievable even for phenotypes with incomplete genotype-to-phenotype mapping, and applicable for indivi...
www.nature.com
David Gokhman
@david-gokhman.bsky.social
· Aug 27
Reposted by David Gokhman
David Gokhman
@david-gokhman.bsky.social
· Jul 26
David Gokhman
@david-gokhman.bsky.social
· Jul 26
David Gokhman
@david-gokhman.bsky.social
· Jul 26
David Gokhman
@david-gokhman.bsky.social
· Jul 26
David Gokhman
@david-gokhman.bsky.social
· Jul 26
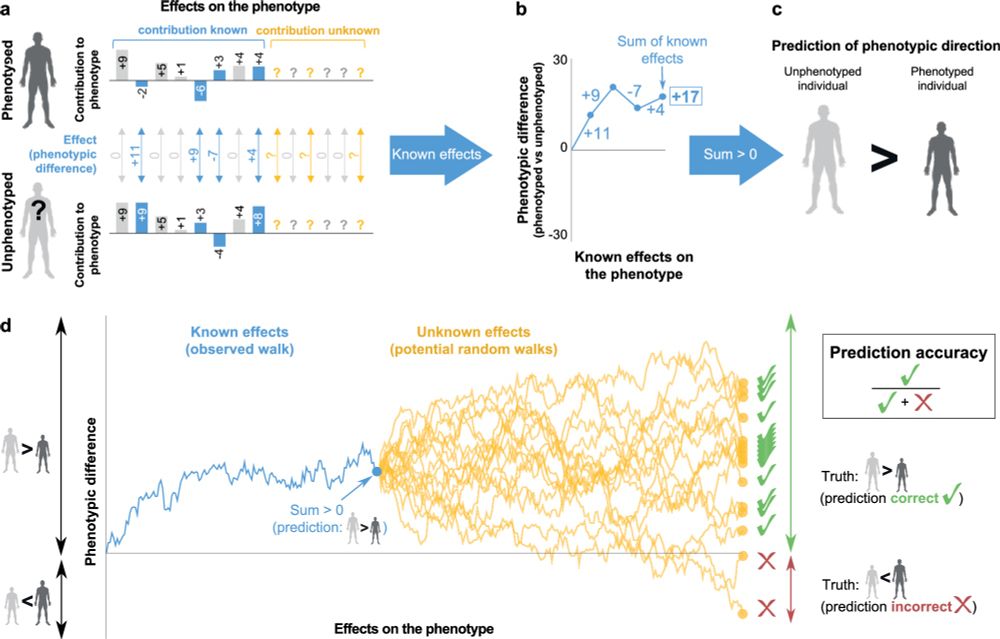
Predicting the direction of phenotypic difference
Nature Communications - Here authors reveal a method to predict key information on phenotypes - their direction. This is achievable even for phenotypes with incomplete genotype-to-phenotype...
rdcu.be
David Gokhman
@david-gokhman.bsky.social
· Jun 20
David Gokhman
@david-gokhman.bsky.social
· Jun 20
David Gokhman
@david-gokhman.bsky.social
· Jun 18
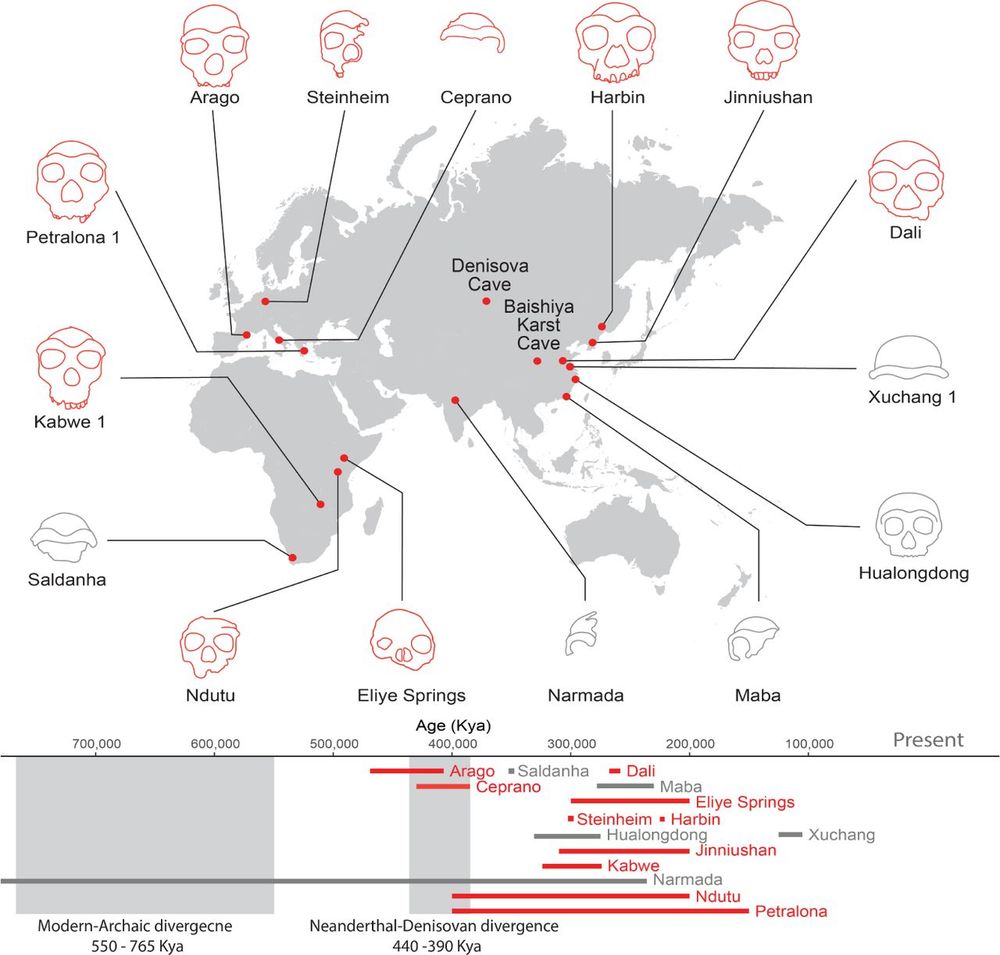
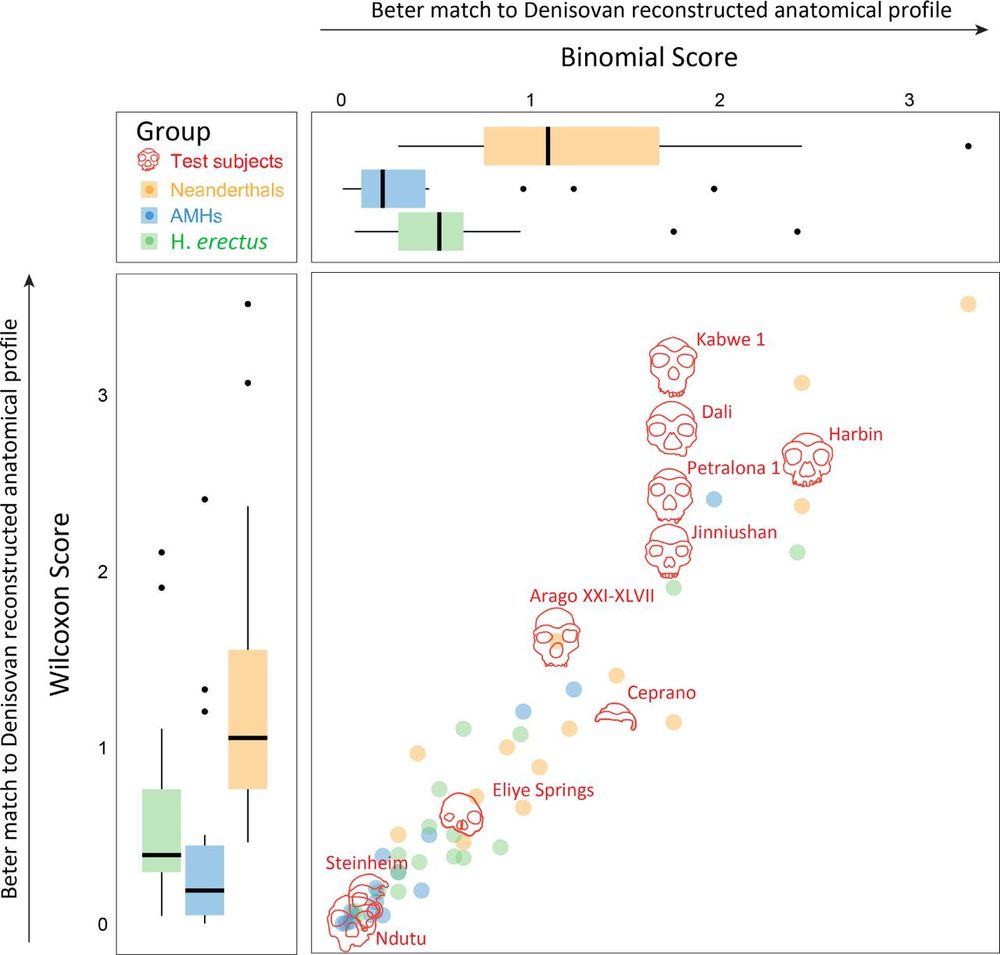
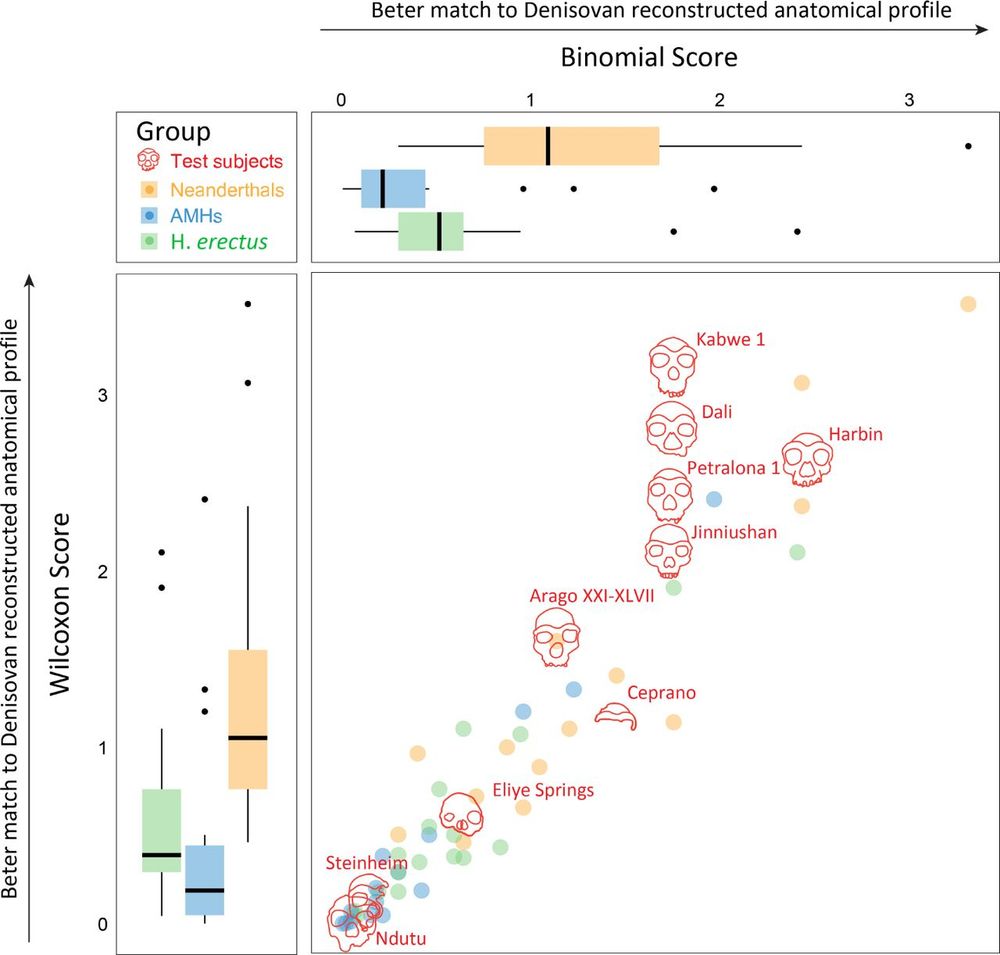
The proteome of the late Middle Pleistocene Harbin individual
Denisovans are a hominin group primarily known through genomes or proteins, but the precise morphological features of Denisovans remain elusive due to the fragmentary nature of discovered fossils. Her...
www.science.org
David Gokhman
@david-gokhman.bsky.social
· Jun 18
David Gokhman
@david-gokhman.bsky.social
· Apr 18