Gene regulation and enhancer design 🧬
Missed the talk?
Here's the preprint: https://www.biorxiv.org/content/10.1101/2025.10.13.682018v1


Missed the talk?
Here's the preprint: https://www.biorxiv.org/content/10.1101/2025.10.13.682018v1

https://tinyurl.com/53nc9vnd
If you're interested in applying AI to biology, take a look at the PhD projects across our labs.
🗓️ Deadline: June 22nd

https://tinyurl.com/53nc9vnd
If you're interested in applying AI to biology, take a look at the PhD projects across our labs.
🗓️ Deadline: June 22nd

www.biorxiv.org/content/10.1...
www.biorxiv.org/content/10.1...

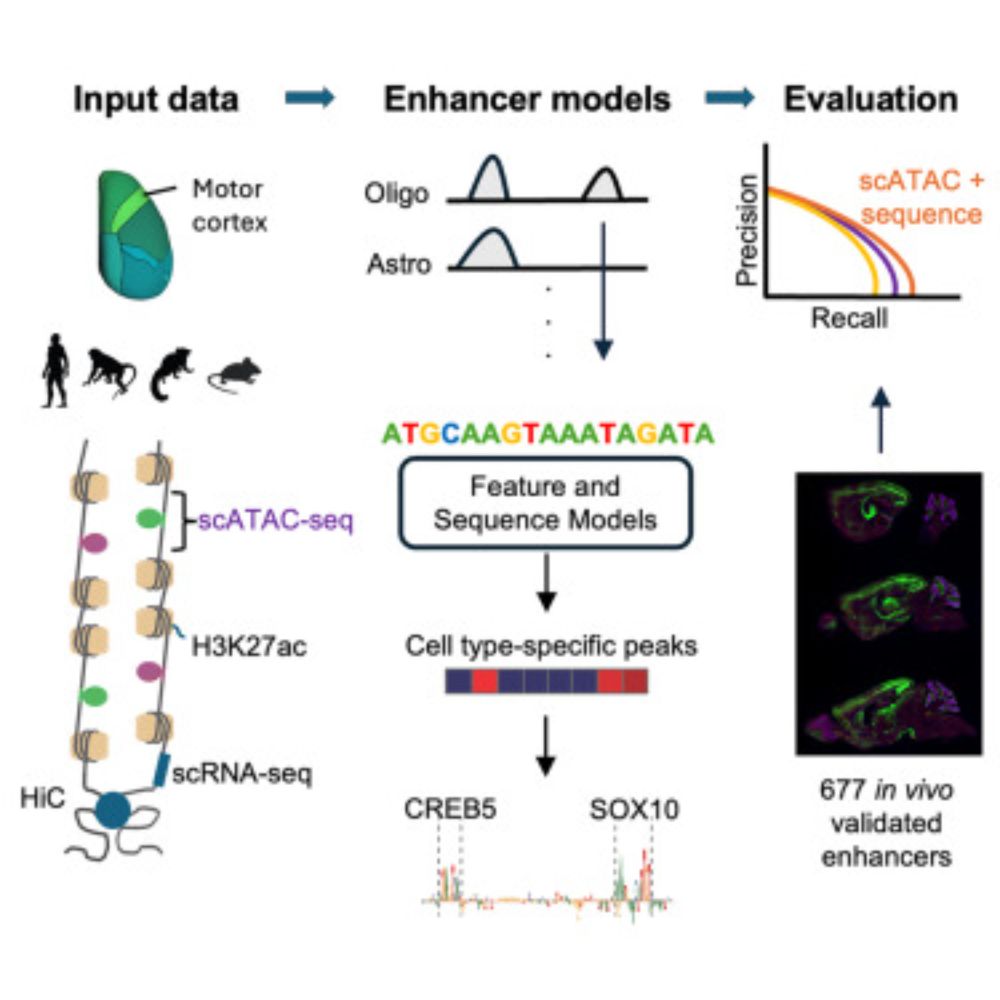
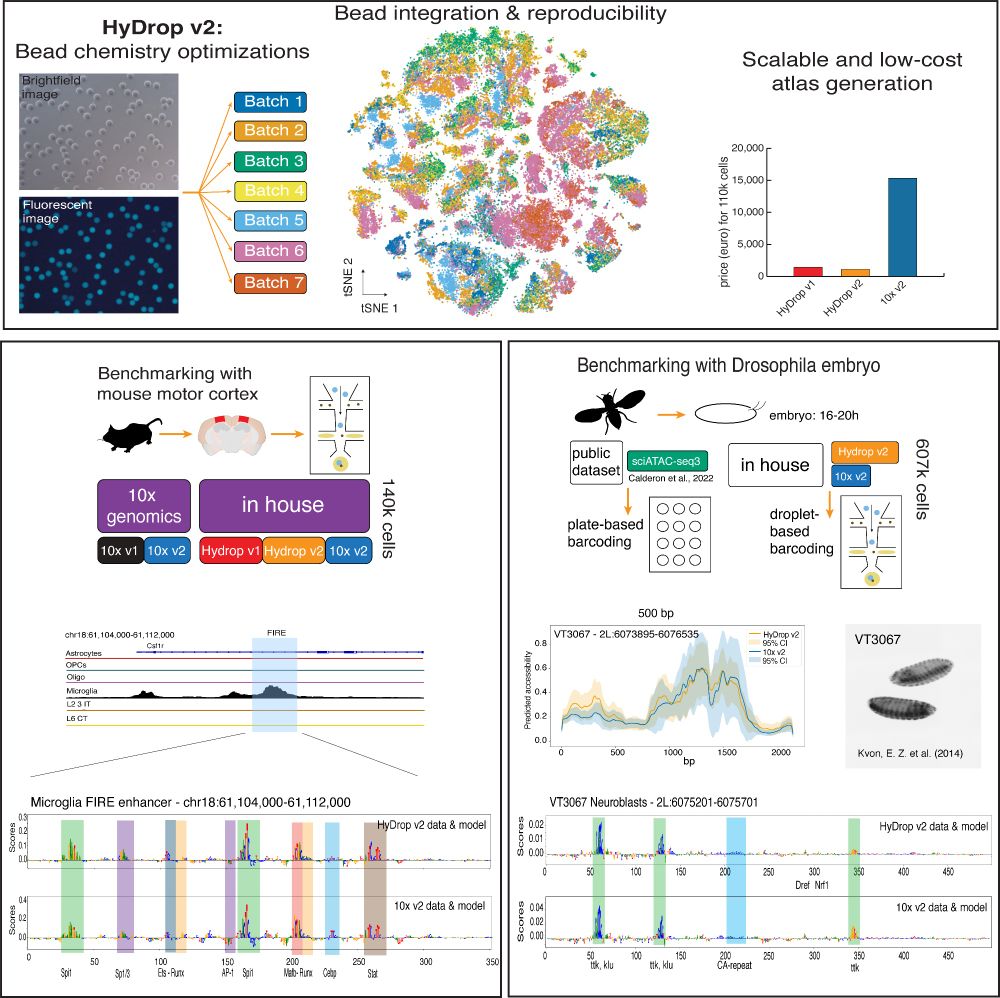
1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...

1) CREsted: to train sequence-to-function deep learning models on scATAC-seq atlases, and use them to decipher enhancer logic and design synthetic enhancers. This has been a wonderful lab-wide collaborative effort. www.biorxiv.org/content/10.1...


