Daan C. Swarts
@dcswarts.bsky.social
210 followers
76 following
36 posts
Group leader at Wageningen University & Research, NL | Prokaryotic immune systems | Views are my own
Posts
Media
Videos
Starter Packs
Daan C. Swarts
@dcswarts.bsky.social
· Aug 28
Daan C. Swarts
@dcswarts.bsky.social
· Aug 28
Daan C. Swarts
@dcswarts.bsky.social
· Jun 16
Daan C. Swarts
@dcswarts.bsky.social
· May 14
Daan C. Swarts
@dcswarts.bsky.social
· May 13
Daan C. Swarts
@dcswarts.bsky.social
· May 13
Daan C. Swarts
@dcswarts.bsky.social
· May 13
Daan C. Swarts
@dcswarts.bsky.social
· May 13
Daan C. Swarts
@dcswarts.bsky.social
· Jan 23
Daan C. Swarts
@dcswarts.bsky.social
· Jan 7
Daan C. Swarts
@dcswarts.bsky.social
· Jan 7

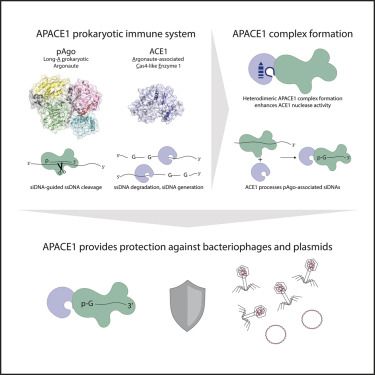
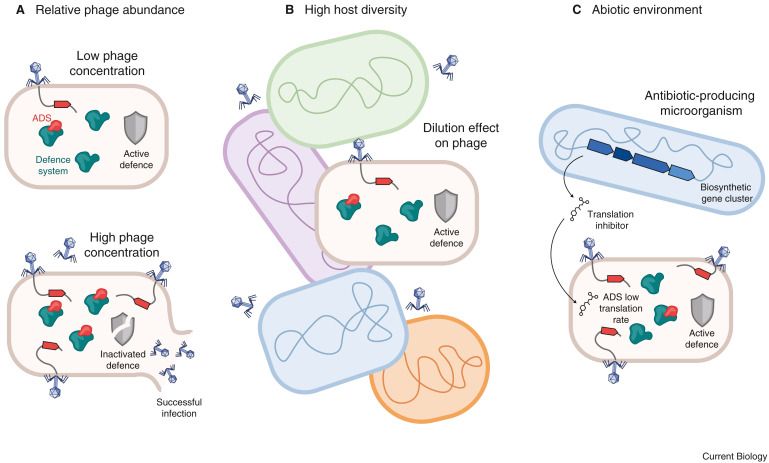
Distribution of bacterial DNA repair proteins and their co-occurrence with immune systems
Bacteria encode various DNA repair pathways to maintain genome integrity. However, the high degree of homology between DNA repair proteins or their do…
www.sciencedirect.com
Daan C. Swarts
@dcswarts.bsky.social
· Jan 7
Daan C. Swarts
@dcswarts.bsky.social
· Jan 7
Daan C. Swarts
@dcswarts.bsky.social
· Jan 7