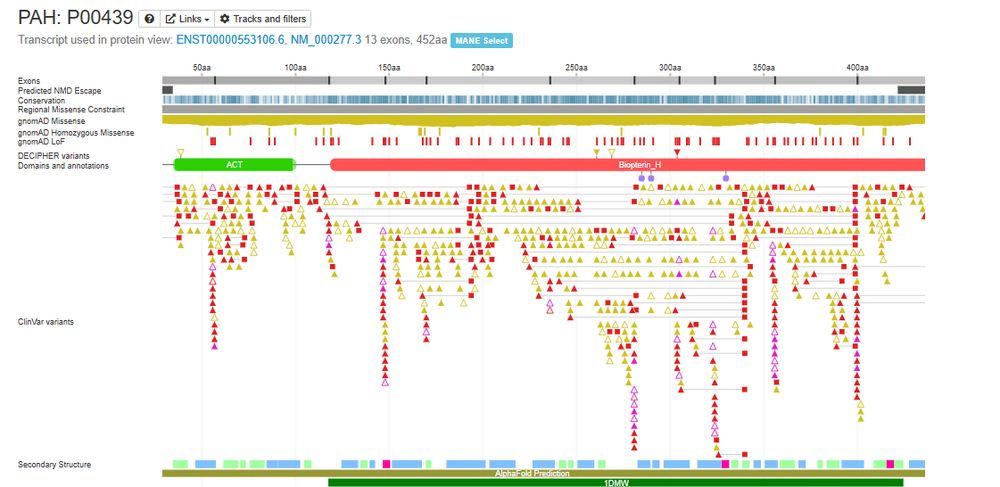
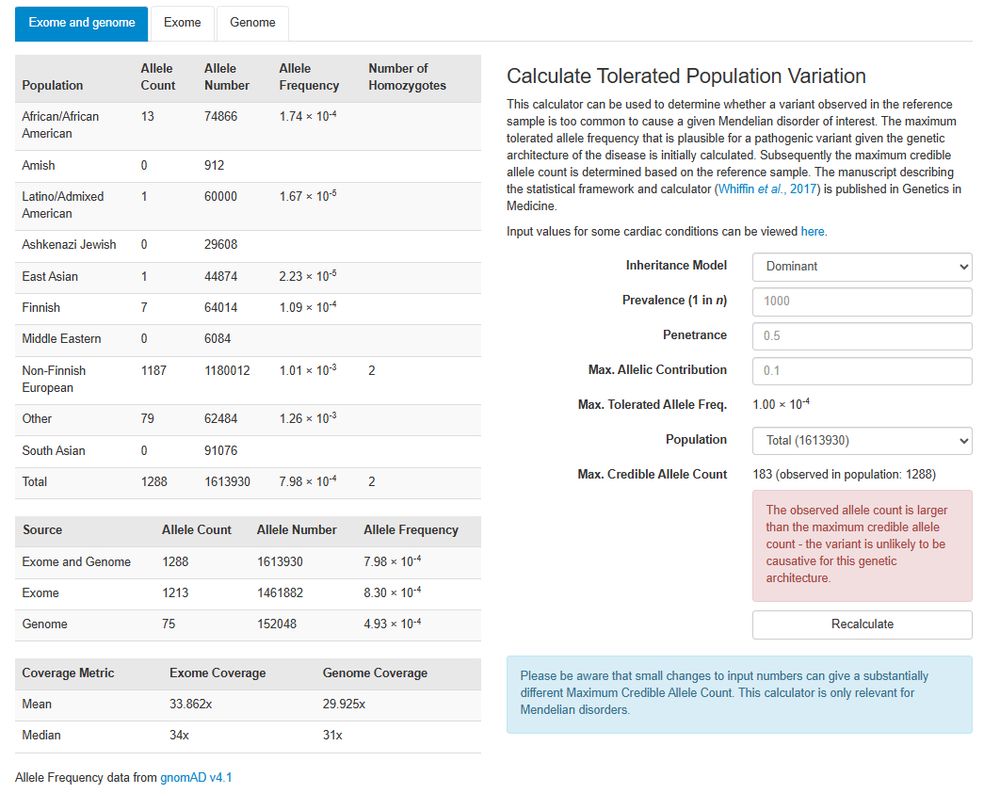
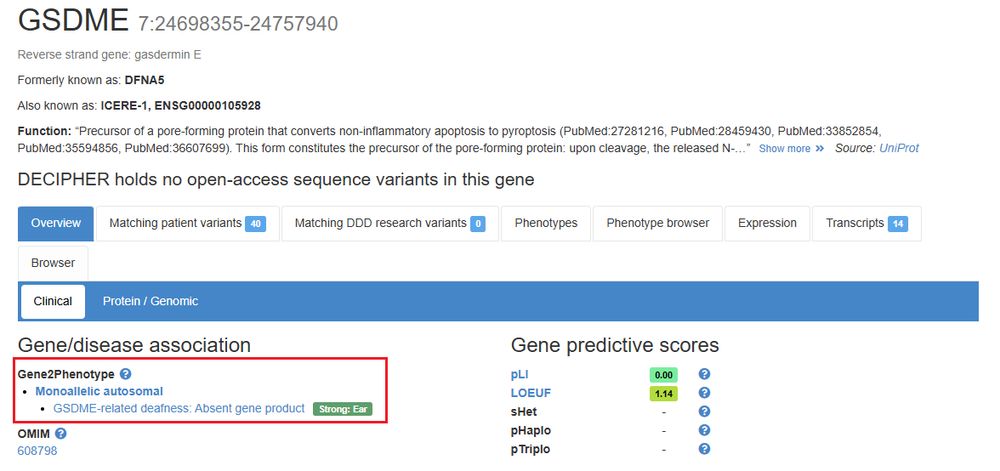
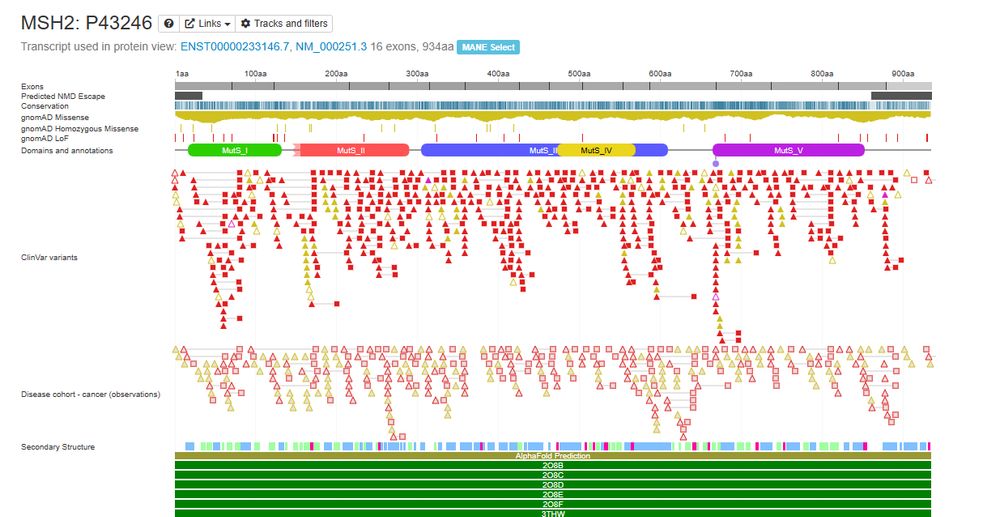
Posts
Media
Videos
Starter Packs
Reposted
Reposted
Reposted
Reposted
Reposted
Reposted
James Fasham
@jamesfasham.bsky.social
· May 25
Reposted