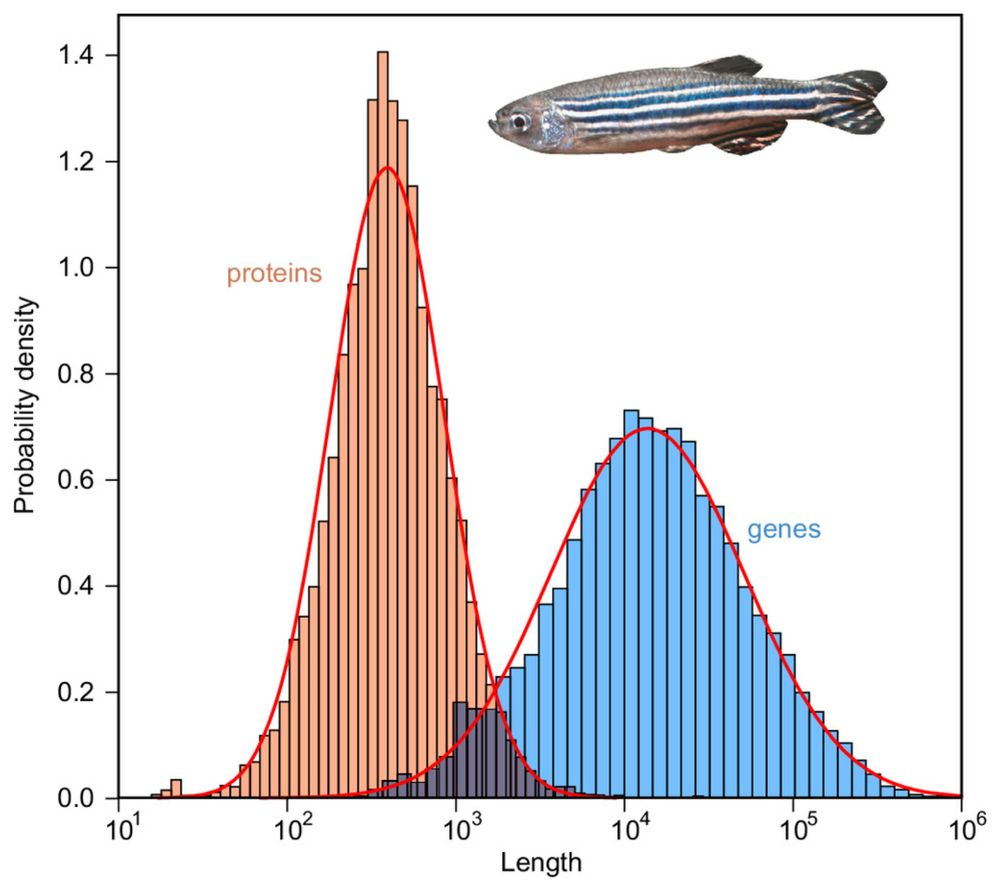
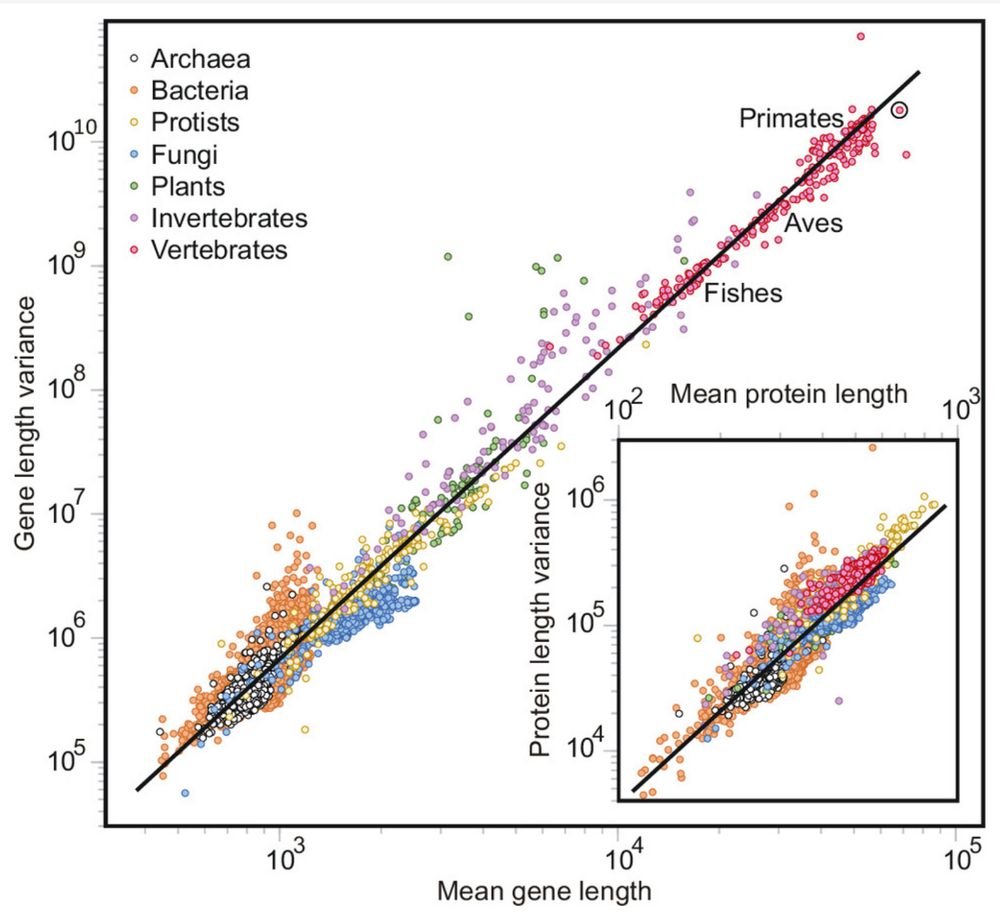
Enrique M. Muro
@emmuro.bsky.social
7 followers
6 following
1 posts
Computational biologist at the Johannes Gutenberg University in Mainz, Germany
https://cbdm-01.zdv.uni-mainz.de/~muro/
https://github.com/emuro
Posts
Media
Videos
Starter Packs
Reposted by Enrique M. Muro
Reposted by Enrique M. Muro
Reposted by Enrique M. Muro