Enrico Orsi
@eorsi.bsky.social
470 followers
37 following
48 posts
Bolognese 🇮🇹 in Copenhagen 🇩🇰
I engineer and optimize the metabolism of nonmodel C1-trophic bacteria for novel biotechnological applications
Metabolic Engineering | Green Economy | CO2 valorization | DTU Biosustain
Posts
Media
Videos
Starter Packs
Enrico Orsi
@eorsi.bsky.social
· Jul 10

🚨 We're hiring a #Postdoc in #Bioprocess #Engineering! | Enrico Orsi
🚨 We're hiring a #Postdoc in #Bioprocess #Engineering! 🌱
📍 Location: DTU Biosustain
📅 Start: January 2026 | 🕐 Duration: 1 year (with potential extension)
Are you a recent PhD graduate excited b...
www.linkedin.com
Enrico Orsi
@eorsi.bsky.social
· Jul 10
Reposted by Enrico Orsi
Enrico Orsi
@eorsi.bsky.social
· May 26

Designed bacteria can detect specific molecules
Researchers have developed a computer-guided method for designing and engineering bacteria that can detect the presence of specific molecules in the environment. This method will advance the use...
www.sciencenews.dk
Reposted by Enrico Orsi
Reposted by Enrico Orsi
Enrico Orsi
@eorsi.bsky.social
· Mar 14
Enrico Orsi
@eorsi.bsky.social
· Mar 14
Enrico Orsi
@eorsi.bsky.social
· Mar 14

Streamlined and efficient genome editing in Cupriavidus necator H16 using an optimised SIBR-Cas system
This study developed two simple, efficient, and rapid genome editing tools termed
Self-splicing Intron-Based Riboswitch-Cas9 (SIBR-Cas9) and SIBR2.0-Cas12a, for editing
the genome of the industrially ...
www.cell.com
Reposted by Enrico Orsi
Qian Cheng
@qchengx1e3.bsky.social
· Mar 12
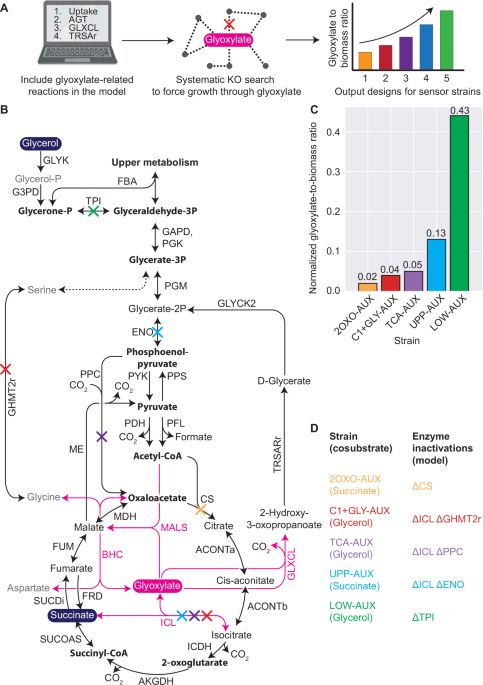
Computation-aided designs enable developing auxotrophic metabolic sensors for wide-range glyoxylate and glycolate detection - Nature Communications
Auxotrophic metabolic sensors (AMS) are vital for bioengineering but are often time-consuming to develop. Here, the authors present a workflow for designing versatile AMS, demonstrating their use for ...
www.nature.com
Reposted by Enrico Orsi
Reposted by Enrico Orsi