Eugen Pfeifer
@eugenpfeifer.bsky.social
160 followers
140 following
16 posts
Junior Professor @
MICALIS, Jouy-en-Josas, INRAE (France)
Passionated about gut phages, especially the temperate and episomal ones (phage-plasmids).
Posts
Media
Videos
Starter Packs
Reposted by Eugen Pfeifer
Eduardo Rocha
@epcrocha.bsky.social
· Sep 3
Reposted by Eugen Pfeifer
Chloé Feltin
@chloe-feltin.bsky.social
· Aug 11

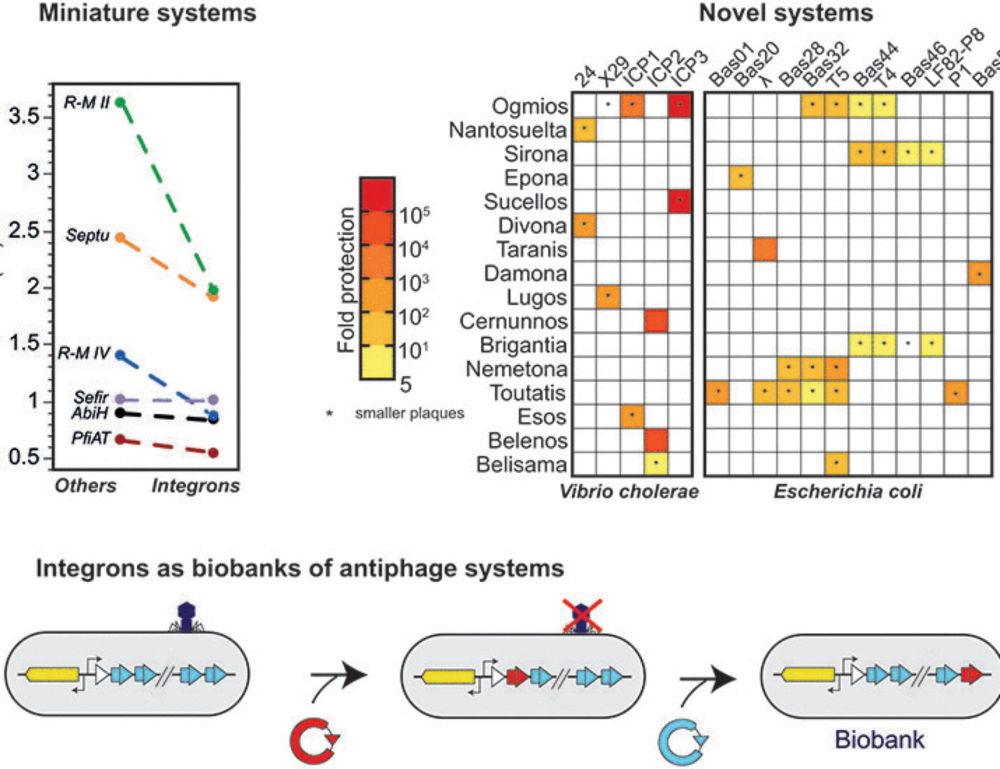
Ecological ubiquity and phylogeny drive nestedness in phages–bacteria networks and shape the bacterial defensome
Identifying the ecological and evolutionary factors that shape phage–bacterial interactions is key to understanding their dynamics in microbial communities. Yet, such interactions remain poorly charac...
www.biorxiv.org
Reposted by Eugen Pfeifer
Eloi Littner
@eloilittner.bsky.social
· May 9
Eugen Pfeifer
@eugenpfeifer.bsky.social
· Jul 28
Eugen Pfeifer
@eugenpfeifer.bsky.social
· Jul 28
Reposted by Eugen Pfeifer
Eugen Pfeifer
@eugenpfeifer.bsky.social
· Feb 10

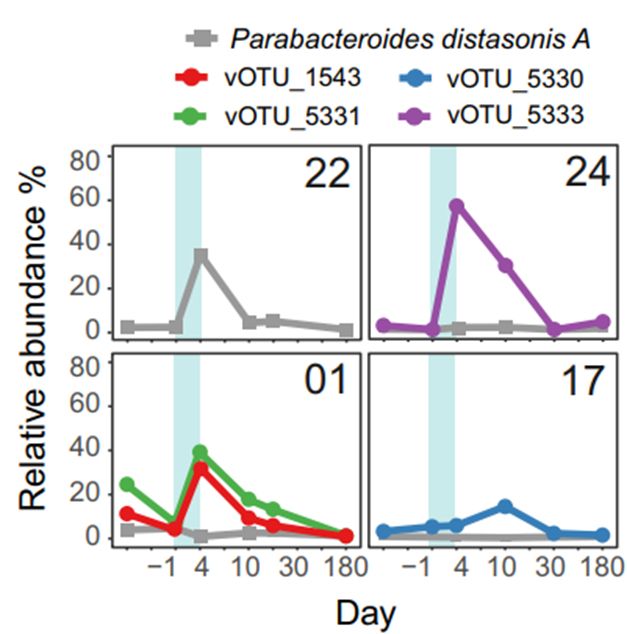
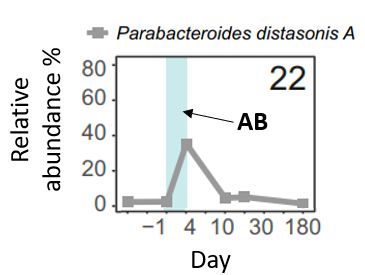
Third-generation cephalosporin antibiotics induce phage bursts in the human gut microbiome.
The use of antibiotics disrupts the gut microbiota, potentially leading to long-term health issues and the spread of resistance. To investigate the impact of antibiotics on phage populations, we follo...
doi.org
Reposted by Eugen Pfeifer
Eduardo Rocha
@epcrocha.bsky.social
· Jan 1