Färkkilä Laboratory
@farkkilab.bsky.social
300 followers
650 following
28 posts
The Systems Medicine of Tumor Microenvironment Lab explores the dynamics of the tumor microenvironment through systems oncology to discover novel biomarkers
Lead by @afarkkila.bsky.social
🌍📍 University of Helsinki, Finland
🔗 https://farkkilab.org/
Posts
Media
Videos
Starter Packs
Pinned
Anniina Färkkilä
@afarkkila.bsky.social
· Dec 10

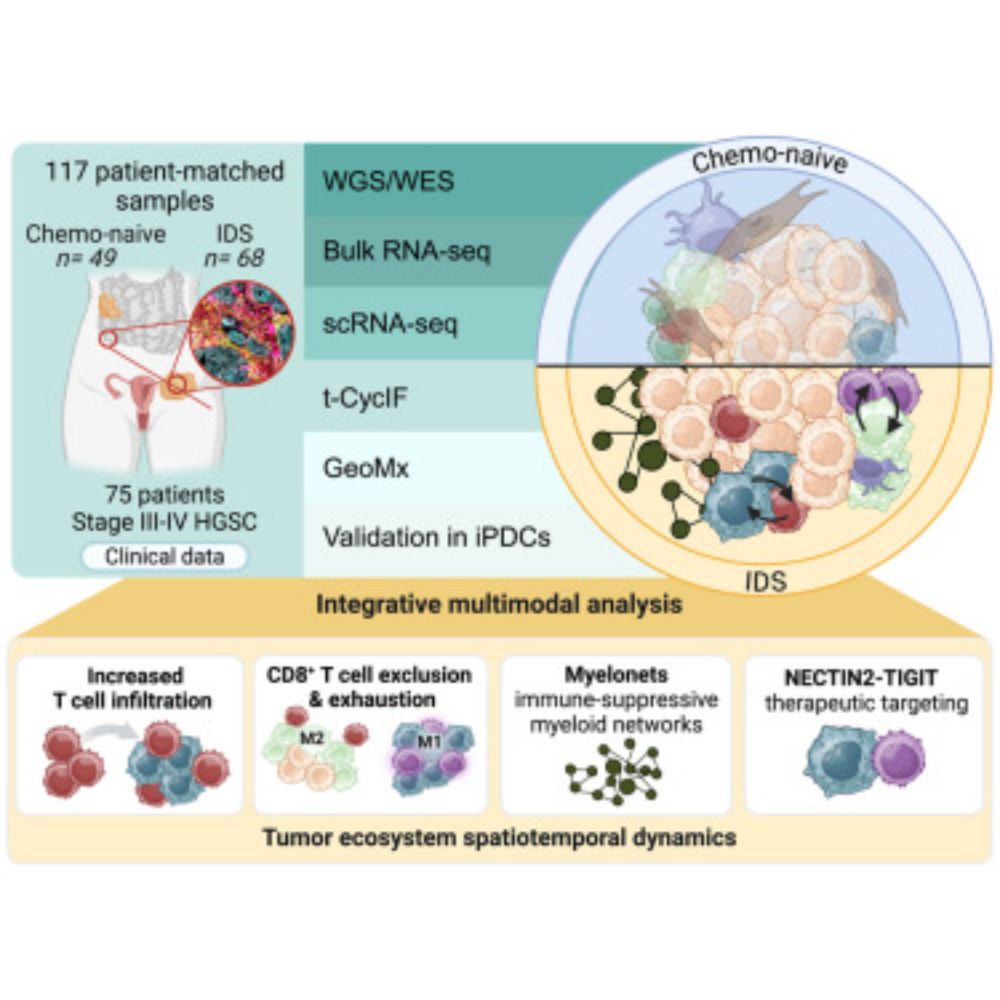
New insights into harnessing the immune system to combat ovarian cancer | University of Helsinki
Researchers have discovered how chemotherapy can change tumors, making them more vulnerable to new types of treatments. These findings could lead to personalized therapies that target the right patien...
www.helsinki.fi