Fengchao
@fcyucn.bsky.social
1K followers
290 following
39 posts
Research Investigator at U of M. Interested in proteomics, etc. Developer of FragPipe, MSFragger, IonQuant, etc.
Posts
Media
Videos
Starter Packs
Reposted by Fengchao
Reposted by Fengchao
Alexey Nesvizhskii
@nesvilab.bsky.social
· Aug 20

Nucleoside diphosphate kinase A (NME1) catalyses its own oligophosphorylation - Nature Chemistry
Our understanding of how post-translational modification—protein phosphorylation—impacts the complexity of eukaryotic signalling pathways is continuously expanding. Now, protein oligophosphorylation h...
www.nature.com
Fengchao
@fcyucn.bsky.social
· Aug 14
Reposted by Fengchao
Reposted by Fengchao
Reposted by Fengchao
Reposted by Fengchao
Alexey Nesvizhskii
@nesvilab.bsky.social
· May 23
Kusterlab
@kusterlab.bsky.social
· May 22
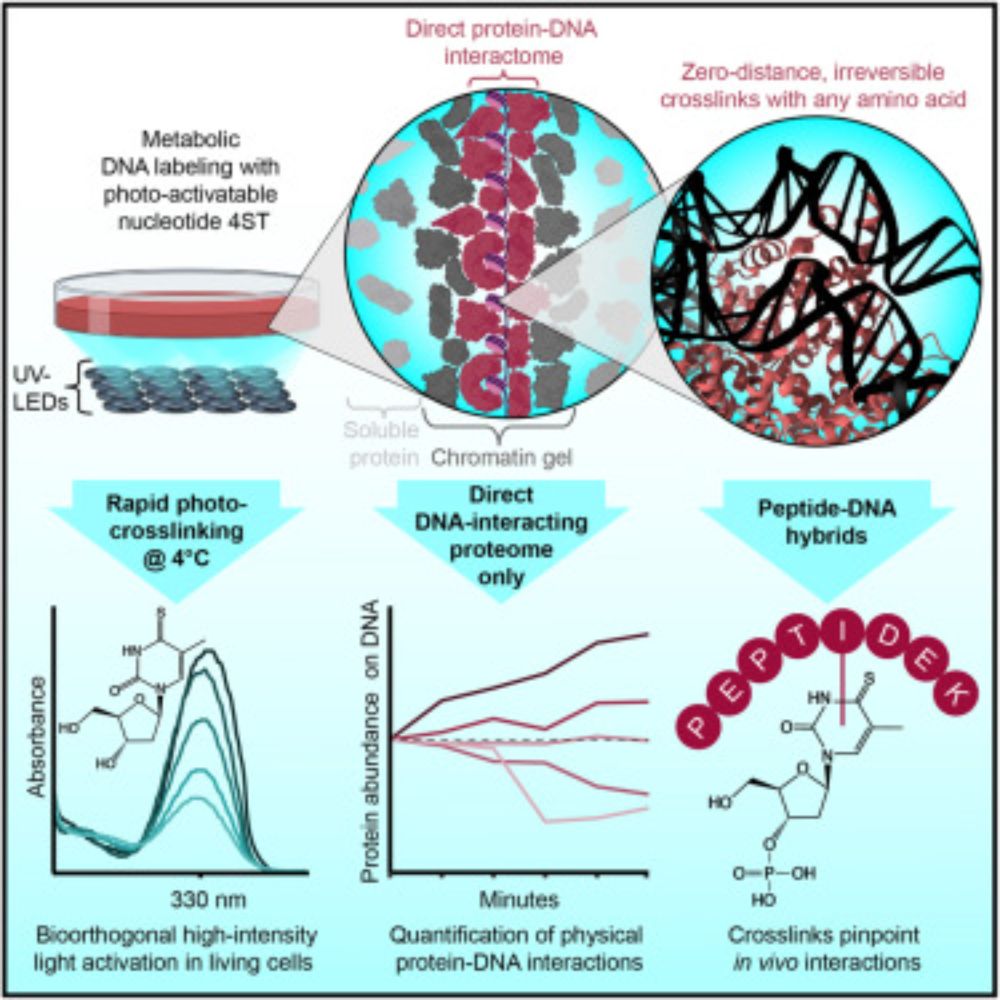
The human proteome with direct physical access to DNA
Zero-distance photo-crosslinking reveals direct protein-DNA interactions in living
cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale
of minutes with single-amino-aci...
www.cell.com
Reposted by Fengchao
Alexey Nesvizhskii
@nesvilab.bsky.social
· May 15
Reposted by Fengchao
Qian Cheng
@qchengx1e3.bsky.social
· Apr 15
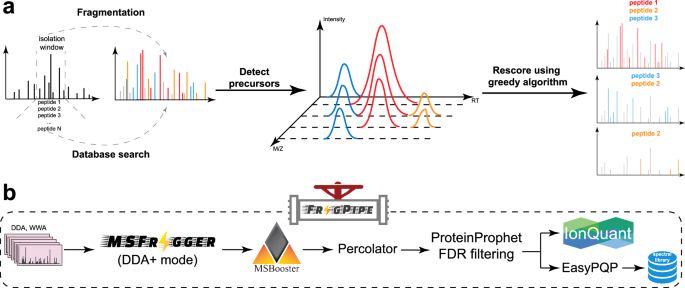
MSFragger-DDA+ enhances peptide identification sensitivity with full isolation window search - Nature Communications
Proteomics often misses co-fragmented peptides in DDA data. Here, the authors introduce MSFragger-DDA+, a database search tool that enhances peptide identification by searching the full isolation wind...
www.nature.com
Fengchao
@fcyucn.bsky.social
· Mar 4
Reposted by Fengchao
Fengchao
@fcyucn.bsky.social
· Jan 4

DIA-Umpire: comprehensive computational framework for data-independent acquisition proteomics - Nature Methods
The computational workflow of DIA-Umpire allows untargeted peptide identificationdirectly from DIA (data-independent acquisition) proteomics data without dependence on a spectral library for data extr...
www.nature.com
Fengchao
@fcyucn.bsky.social
· Jan 4

diaTracer enables spectrum-centric analysis of diaPASEF proteomics data - Nature Communications
Data-independent acquisition advances proteomics quantification. Here, the authors present diaTracer, a spectrum-centric tool for diaPASEF data that supports broad proteomics applications, enabling di...
www.nature.com
Reposted by Fengchao
Fengchao
@fcyucn.bsky.social
· Jan 4

IMTBX and Grppr: Software for Top-Down Proteomics Utilizing Ion Mobility-Mass Spectrometry
Top-down proteomics has emerged as a transformative method for the analysis of protein sequence and post-translational modifications (PTMs). Top-down experiments have historically been performed prima...
doi.org