Posts
Media
Videos
Starter Packs
fowlerlab.org
@fowlerlab.org
· Jul 7

CRyPTIC datasets available through new website
The CRyPTIC project ran from 2016 to 2022 and collected >20,000 clinical samples from patients with tuberculosis. Each sample underwent whole genome sequencing and also was inoculated onto a 96-well plate containing 13 different antibiotics at a range of concentrations. We also collated some existing datasets and the project continued to collect data following the original data freeze in April 2020.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jul 3

Third Dx4LMICs conference
Most of us attended the third Diagnostics for Low- and Middle-Income Countries (Dx4LMICs) conference at Reuben College in Oxford over the last two days. I am a Fellow at Reuben College and helped our President, Professor Lord Lionel Tarassenko and several others, including Dylan Adlard, organise the conference. It was the biggest conference the college has held to date with about 110-120 attendees and we had to close registration a few weeks before and had a waiting list.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jul 3

CRyPTIC datasets available through new website
The CRyPTIC project ran from 2016 to 2022 and collected >20,000 clinical samples from patients with tuberculosis. Each sample underwent whole genome sequencing and also was inoculated onto a 96-well plate containing 13 different antibiotics at a range of concentrations. We also collated some existing datasets and the project continued to collect data following the original data freeze in April 2020.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jul 1
I’m hiring!
Thanks to funding from the OxCoD4TB project, I am hiring a postdoctoral research associate. This consortium pulls together expertise from all over the University, including in diagnostics, data science, drug and vaccine design, preclinical testing and clinical testing and brings it to bear on tuberculosis (TB). The main aims of OxCoD4TB are to establish and validate a new TB therapeutics pipeline and discover and evaluate novel compounds and also drug and vaccine combinations.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jul 1
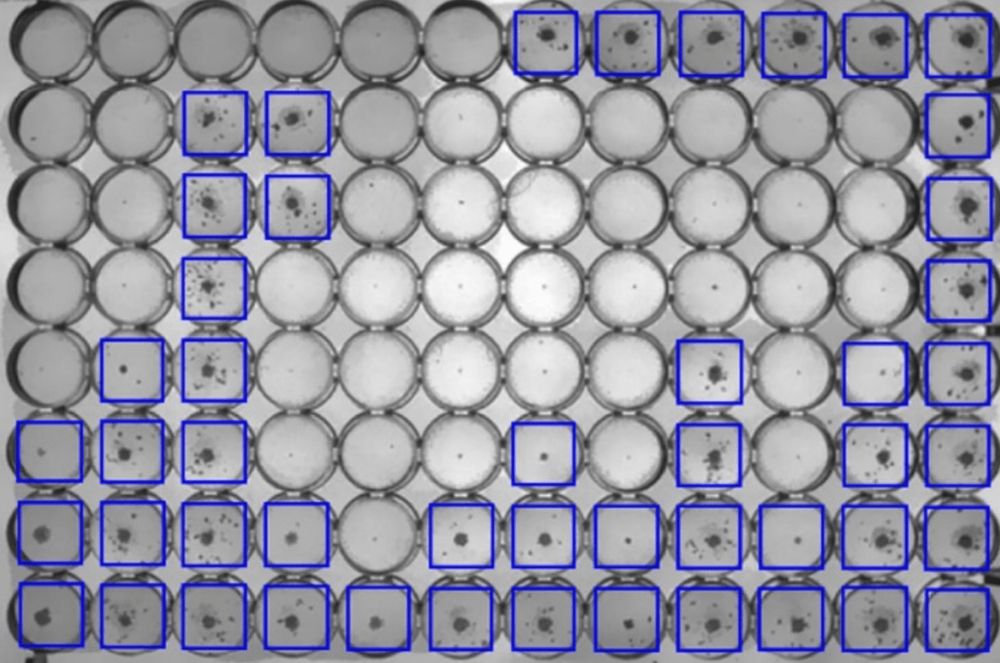
New paper: a deep learning model that reads MICs from images of 96 well plates
Our paper describing how a convolutional neural network model can determine the minimum inhibitory concentrations (MICs) from a photograph of the 96-well plate after two weeks incubation has been published in the Computational and Structural Biology Journal. You can get the model, which is called TMAS, on GitHub here and there is a longer description here.
fowlerlab.org
Reposted
fowlerlab.org
@fowlerlab.org
· Jun 25

ESM Annual Congress 2025
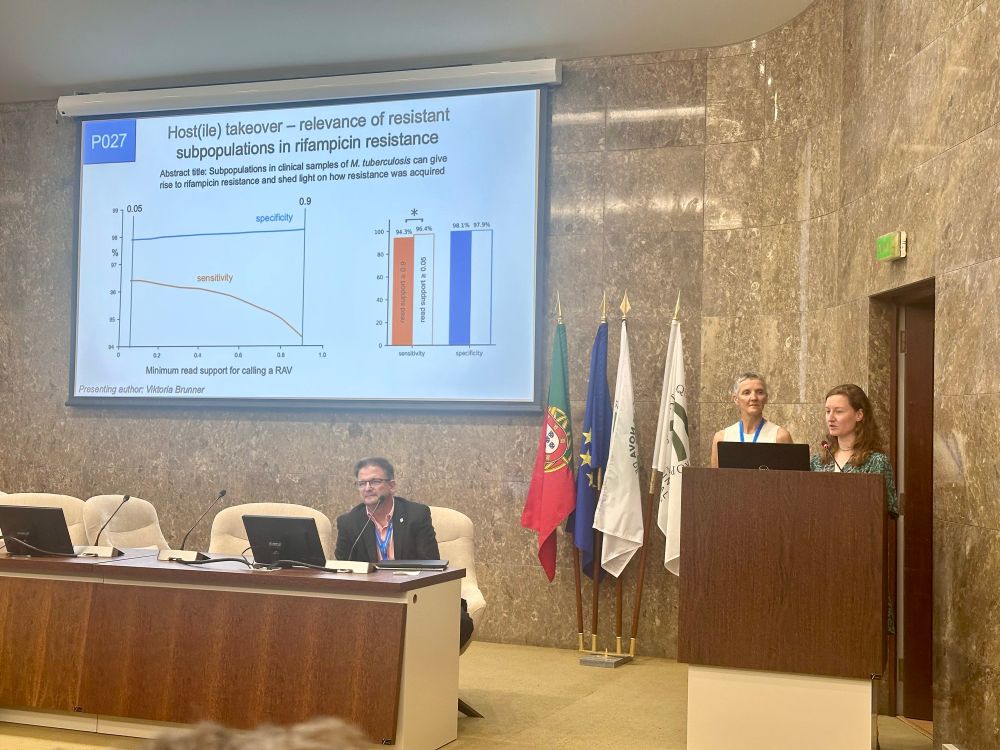
Several of us attended the 45th Annual Congress of the European Society of Mycobacteriology in Lisbon, Portugal. This is probably my favourite scientific meeting -- small, friendly and with plenty of time to talk, including organised social events. Viki Brunner, Dylan Adlard and Philip Fowler all had posters and there were another three posters from the MMM Unit. At the least minute we were offered a short ten minute talk as another speaker could not travel so we gladly accepted the chance and Philip Fowler presented some work analysing 56,306 Mycobacterial samples sequenced by UKHSA between 2016 and 2024. The main aim was to develop and validate an approach that can identify the species present.
fowlerlab.org
Reposted
Reposted
Reposted
Viktoria Brunner
@vbrunner.bsky.social
· Apr 10
fowlerlab.org
@fowlerlab.org
· Apr 10
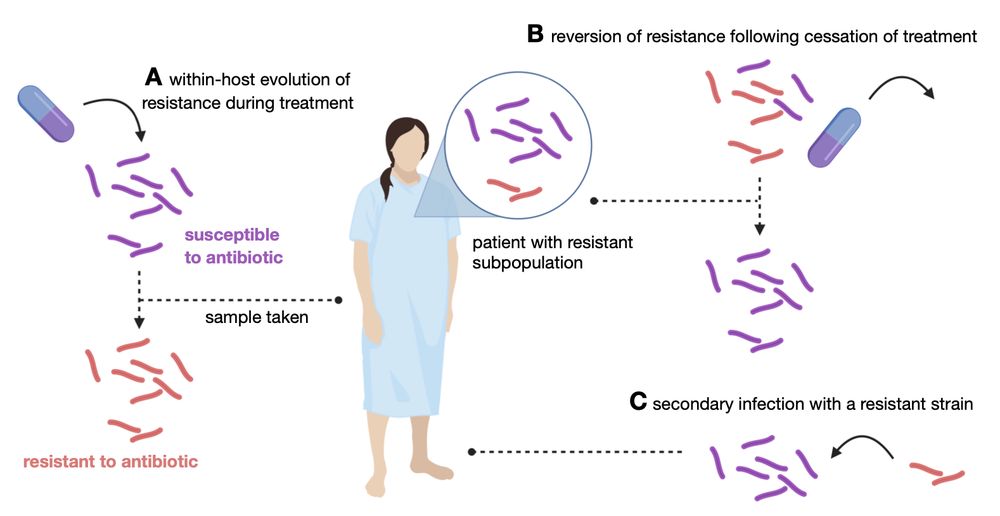
New preprint: looking at rifampicin-resistant subpopulations in clinical samples
Since clinical samples are usually grown in a MGIT tube for a while before some "crumbs" are harvested for DNA extraction, they are metagenomic in the sense that they can and do contain multiple colonies. This means we should expect subpopulations in our analysis but most bioinformatics tools and file formats inherently assume a homogenous sample with a single genome.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Mar 17
New grant: Ox4TB
Very pleased to announce that I am a co-investigator on the recently announced Oxford4TB project that has been funded by the Ineos Oxford Institute for antimicrobial research (IOI). The project will received £5 million over three years and the main aim is to develop new therapies for multi-drug resistant tuberculosis. We will be guiding the selection of targets and leads using our large dataset of >50k clinical tuberculosis samples.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Mar 11
Can medical microbiology become a big data science? Lessons from CRyPTIC
The CRyPTIC project ran from 2017 to around 2022 and in that time collected over 20,000 clinical samples of M. tuberculosis. Each sample underwent whole genome sequencing and phenotypic drug susceptibility testing (pDST); the minimum inhibitory concentrations (MICs) of 13 different antibiotics was tested using a bespoke 96-well broth microdilution plate. The project also aggregated previously published samples which has had pDST data and had undergone WGS.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Feb 23
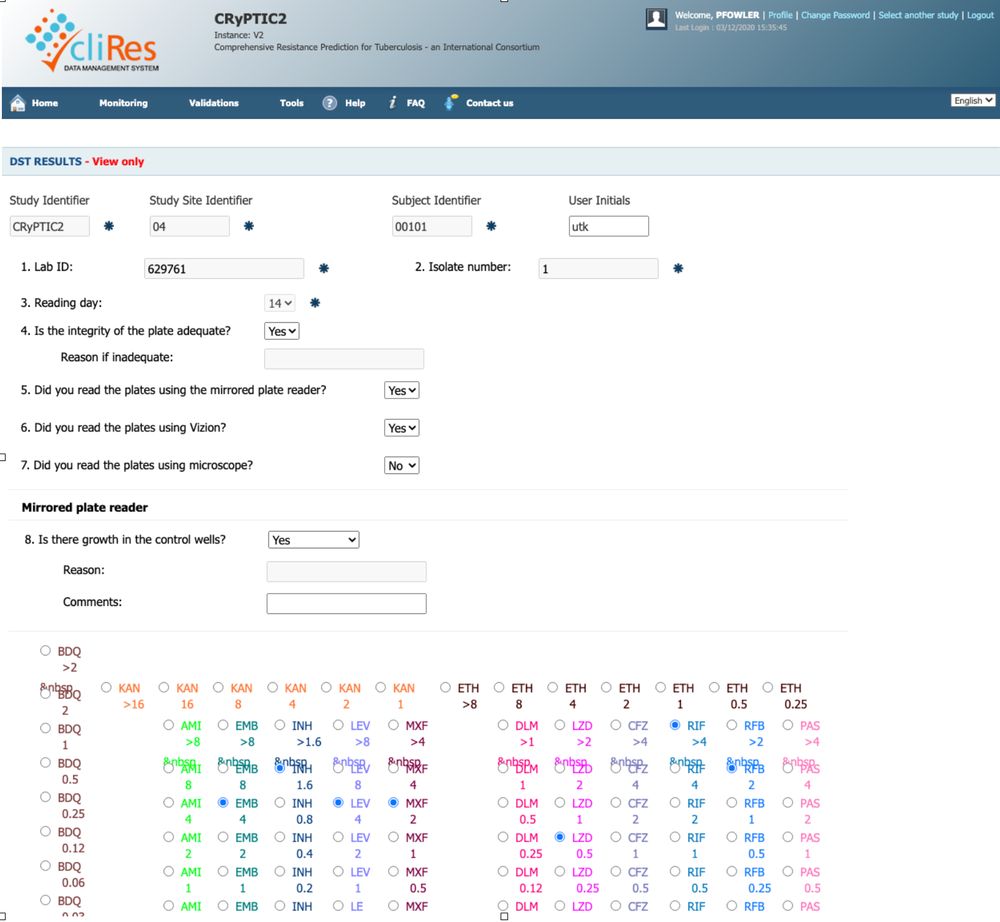
New preprint: a deep learning model that can read 96-well broth micro dilution plates
The CRyPTIC project used bespoke 96-well broth microdilution plates to measure the minimum inhibitory concentrations (MICs) of 13 different antibiotics; to reduce the error in the measurements, photographs of each plate were taken after two weeks incubation and stored. Hence we have available over 20,000 images of M. tuberculosis growing on these plates along with MICs measured using a variety of approaches.
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jan 31
New preprint: automatically building a better bedaquiline catalogue
A catalogue recording whether individual mutations confer resistance or not to specified antibiotics is a necessary component of genetics-based clinical microbiology. Such catalogues need to be not only accurate but also meet a number of minimum requirements if they are to be used widely. Dylan Adlard, who is studying for his PhD, has developed a python package, catomatic, that automatically applies some logic evolved from the…
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jan 28
SARS-CoV-2 pipeline live on EIT Pathogena
Back in the SARS-CoV-2 pandemic we worked closely with ORACLE Corp to build and deploy a bioinformatics pipeline in ORACLE Cloud that processes raw genetic files and infers what the consensus genome is and hence what lineage (e.g. BA.2) it belongs to. The heart of the pipeline is an amplicon-aware variant caller, viridian, that was written by Zamin Iqbal's group and there is a…
fowlerlab.org
fowlerlab.org
@fowlerlab.org
· Jan 28

Updated preprint: A validated cloud-based genomic platform for co-ordinated, expedient global analysis of SARS-CoV-2 genomic epidemiology
In August 2022 seven laboratories across the world uploaded their SARS-CoV-2 genetics files for processing to an online cloud processing platform. Since then the pipeline that is run (the Tiled Amp…
fowlerlab.org