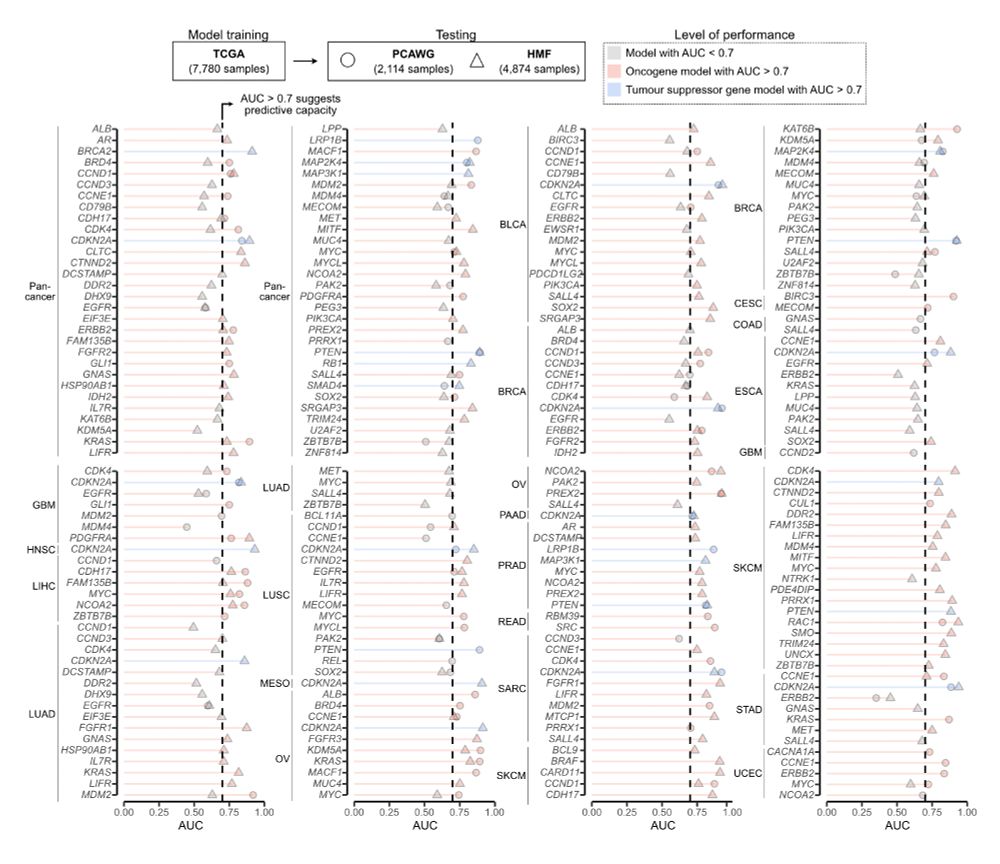
Geoff Macintyre
@gmaci.bsky.social
2.1K followers
1.1K following
35 posts
Group leader in computational oncology at CNIO, Madrid. CSO at Tailor Bio. Chromosomal instability and tumour evolution. www.macintyrelab.org
Posts
Media
Videos
Starter Packs
Geoff Macintyre
@gmaci.bsky.social
· Jun 23
Geoff Macintyre
@gmaci.bsky.social
· Jun 23
Geoff Macintyre
@gmaci.bsky.social
· Jun 23
Geoff Macintyre
@gmaci.bsky.social
· Jun 23
Geoff Macintyre
@gmaci.bsky.social
· Jun 23
Geoff Macintyre
@gmaci.bsky.social
· Jun 23
Geoff Macintyre
@gmaci.bsky.social
· Jun 10
Geoff Macintyre
@gmaci.bsky.social
· Jun 10
Geoff Macintyre
@gmaci.bsky.social
· Jun 10
Geoff Macintyre
@gmaci.bsky.social
· Jun 10
Geoff Macintyre
@gmaci.bsky.social
· Jun 10