Lauren Hemara
@hemara.nz
550 followers
800 following
10 posts
🦠 Postdoc in the Microbiome Manipulation Lab @ University of Toronto
🥝 Previously studying plant pathogen evolution with everyone's favourite P. syringae pathovar - Psa! ✨
🌿 hemara.nz
Posts
Media
Videos
Starter Packs
Reposted by Lauren Hemara
Reposted by Lauren Hemara
Reposted by Lauren Hemara
Reposted by Lauren Hemara
Reposted by Lauren Hemara
Reposted by Lauren Hemara
Michelle Hulin
@michhulin.bsky.social
· Aug 7

From genes to epidemics: Genomic insights into bacterial plant pathogen emergence
Bacterial phytopathogens are major causal agents of newly emerging plant diseases. The roles of both agricultural practices and the alteration of bact…
www.sciencedirect.com
Reposted by Lauren Hemara
Reposted by Lauren Hemara
Kevin Hockett
@hockettlab.bsky.social
· Jul 1

Rapid and sustained differentiation of disease-suppressive phyllosphere microbiomes in tomato following experimental microbiome selection - Environmental Microbiome
Background Microbial-based treatments to protect plants against phytopathogens typically focus on soil-borne disease or the aboveground application of one or a few biocontrol microorganisms. However, ...
tinyurl.com
Reposted by Lauren Hemara
T3SS
@t3sss.bsky.social
· Apr 2
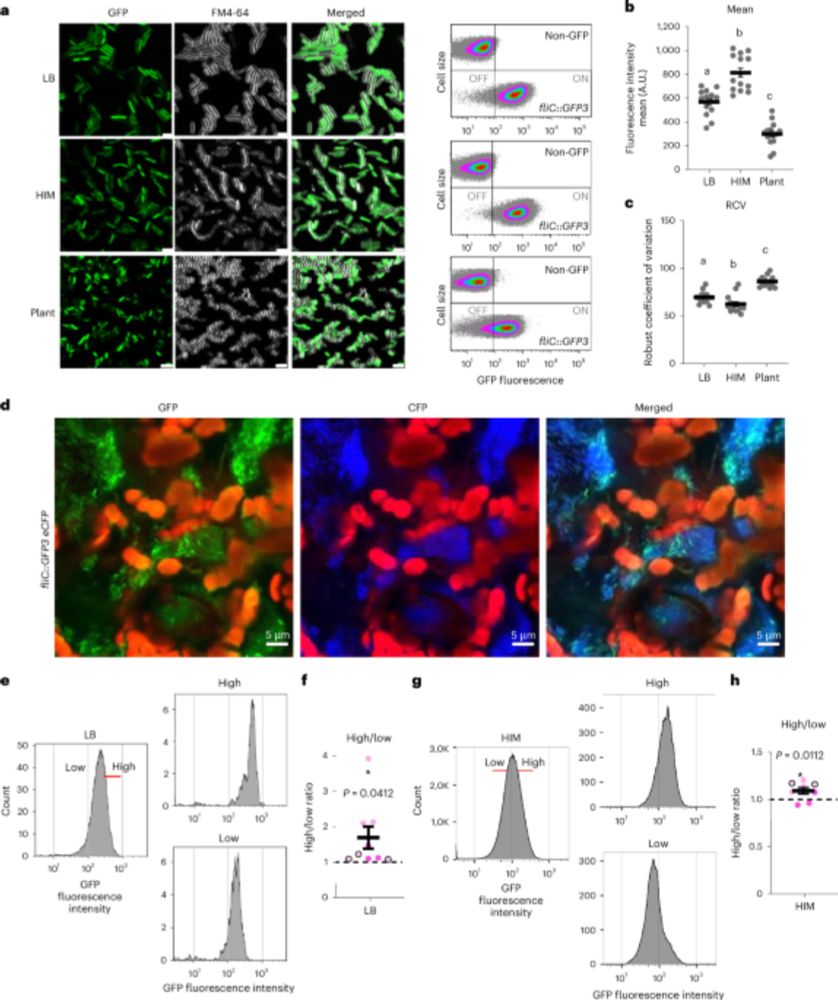
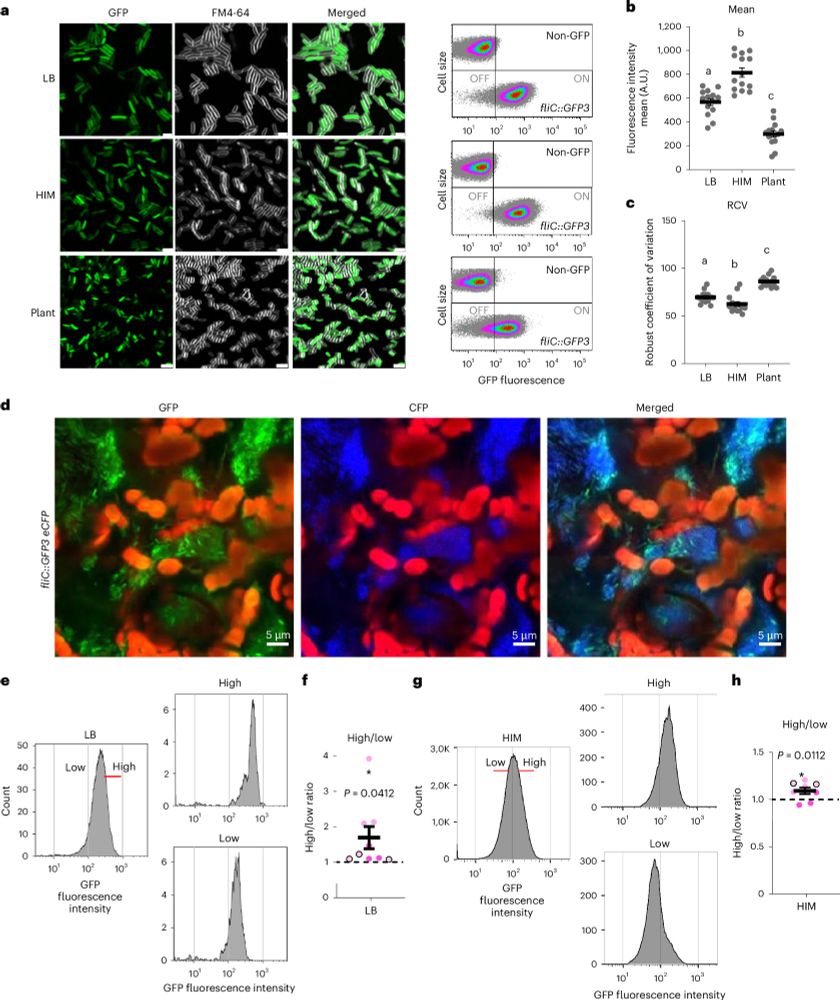
Pseudomonas syringae subpopulations cooperate by coordinating flagellar and type III secretion spatiotemporal dynamics to facilitate plant infection - Nature Microbiology
Single-cell gene expression analysis reveals phenotypic heterogeneity to enable bacterial specialization over the course of plant colonization.
www.nature.com
Reposted by Lauren Hemara
Lauren Hemara
@hemara.nz
· Feb 7

Genomic Biosurveillance of the Kiwifruit Pathogen Pseudomonas syringae pv. actinidiae Biovar 3 Reveals Adaptation to Selective Pressures in New Zealand Orchards
A hopF1c effector loss variant has emerged in commercial kiwifruit orchards, mediated by the introduction of copper resistance elements into the population.
bsppjournals.onlinelibrary.wiley.com
Reposted by Lauren Hemara
Rudolf Schlechter
@rschlec.bsky.social
· Jan 10
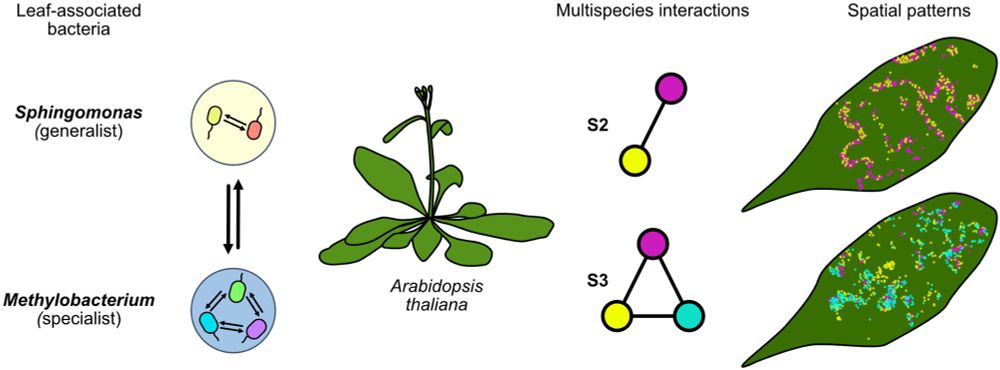
Differential Responses of Methylobacterium and Sphingomonas Species to Multispecies Interactions in the Phyllosphere
Leaf-associated bacterial populations of Methylobacterium and Sphingomonas in two- and three-member synthetic communities on Arabidopsis thaliana display distinct spatial structures and population dy....
enviromicro-journals.onlinelibrary.wiley.com
Reposted by Lauren Hemara
Lauren Hemara
@hemara.nz
· Oct 25
Lauren Hemara
@hemara.nz
· Oct 25

Genomic biosurveillance of the kiwifruit pathogen Pseudomonas syringae pv. actinidiae biovar 3 reveals adaptation to selective pressures in New Zealand orchards
In the late 2000s, a pandemic of Pseudomonas syringae pv. actinidiae biovar 3 (Psa3) devastated kiwifruit orchards growing susceptible yellow-fleshed cultivars. New Zealand's kiwifruit industry has si...
www.biorxiv.org