Jeff Vierstra
@jeffvierstra.bsky.social
370 followers
400 following
130 posts
Senior Investigator @ Altius Institute for Biomedical Sciences. Research: High-resolution mapping of chromatin structure & function. Fun: Mountain shenanigans and skiing turns all year. Seattle, USA/Patagonia Chilena (🇺🇸🇨🇱). http://vierstra.org
Posts
Media
Videos
Starter Packs
Pinned
Reposted by Jeff Vierstra
Jeff Vierstra
@jeffvierstra.bsky.social
· Aug 13
Jeff Vierstra
@jeffvierstra.bsky.social
· Aug 13
Reposted by Jeff Vierstra
Jeff Vierstra
@jeffvierstra.bsky.social
· Jul 27
Jeff Vierstra
@jeffvierstra.bsky.social
· May 24
Jakob Trendel
@jtrend.bsky.social
· May 23
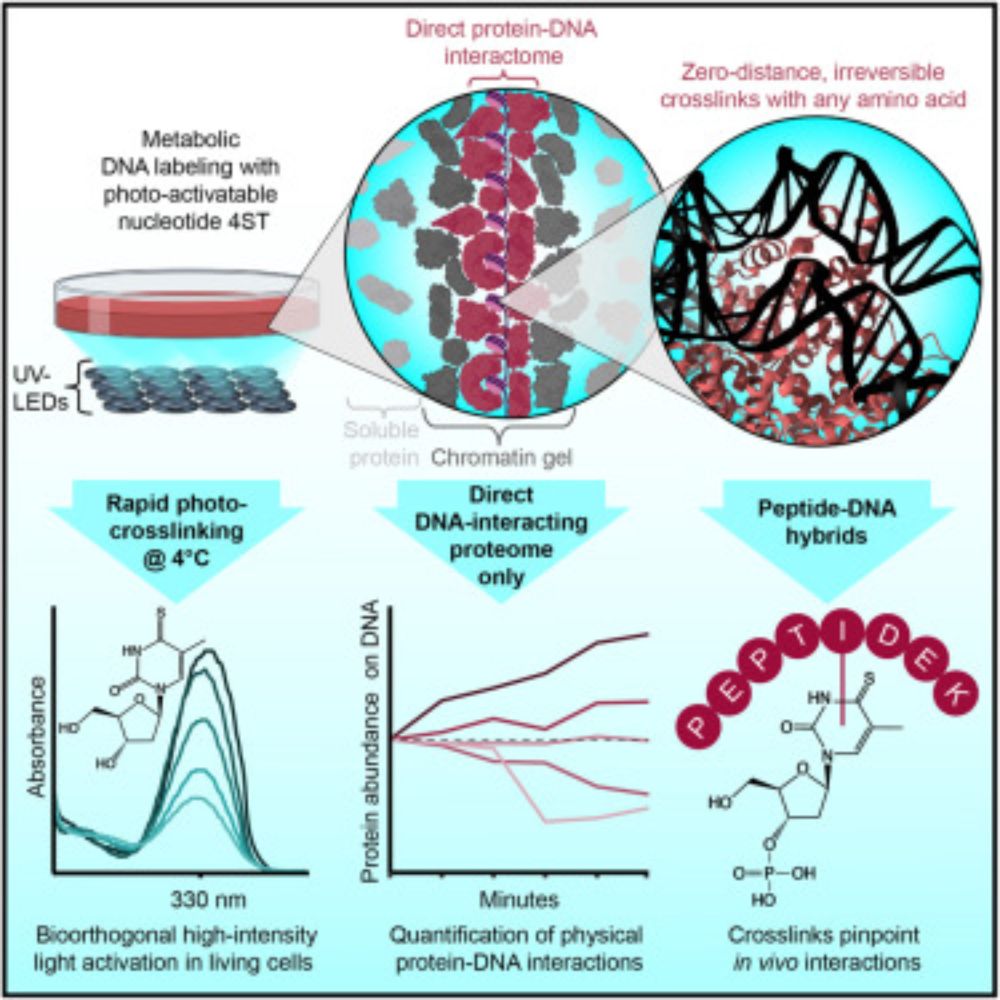
The human proteome with direct physical access to DNA
Zero-distance photo-crosslinking reveals direct protein-DNA interactions in living
cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale
of minutes with single-amino-aci...
www.cell.com
Jeff Vierstra
@jeffvierstra.bsky.social
· May 23