Jenna Elliott
@jennaelliott.bsky.social
62 followers
64 following
12 posts
Biology-inspired physics | Information processing | PhD student in the Erzberger group at EMBL | she/her
Posts
Media
Videos
Starter Packs
Reposted by Jenna Elliott
Jenna Elliott
@jennaelliott.bsky.social
· Jun 20
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18

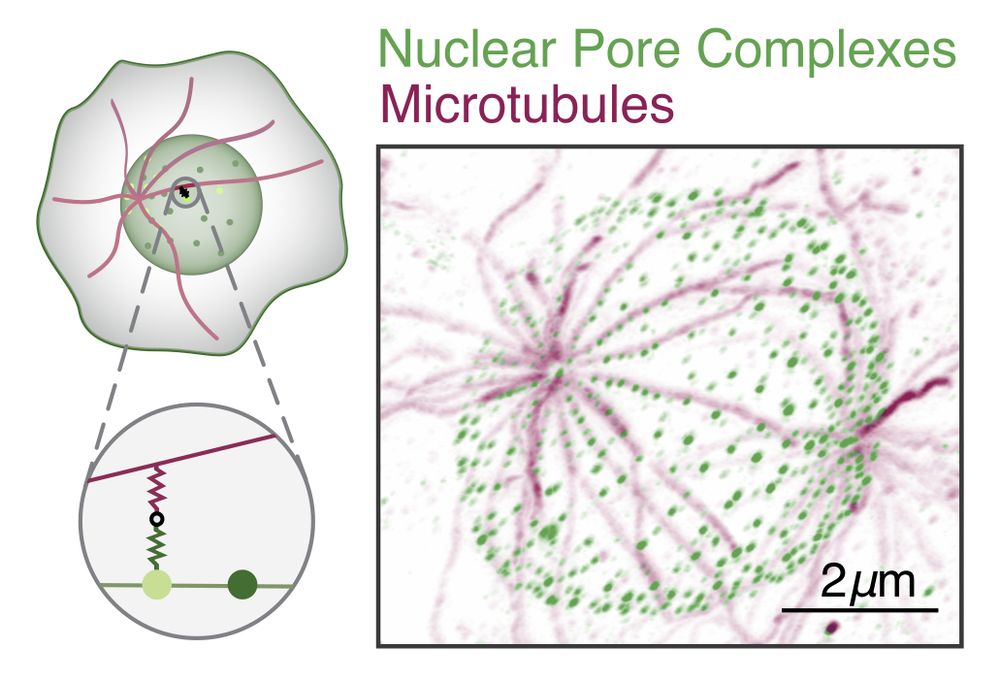
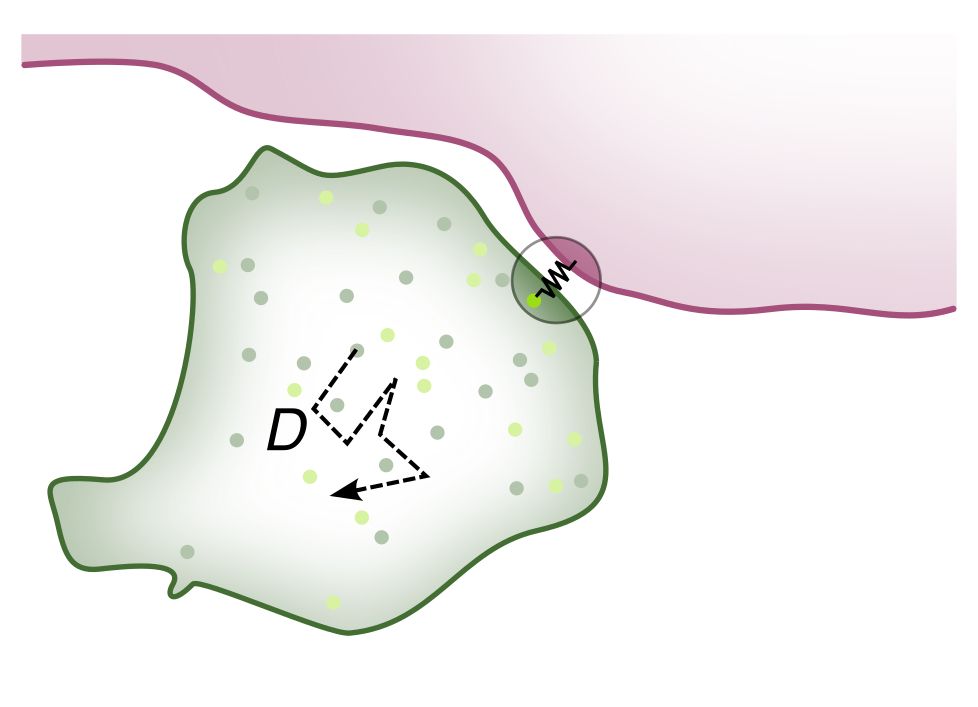
Repulsive particle interactions enable selective information processing at cellular interfaces
Living systems relay information across membrane interfaces to coordinate compartment functions. We identify a physical mechanism for selective information transmission that arises from the sigmoidal ...
arxiv.org
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18
Jenna Elliott
@jennaelliott.bsky.social
· Jun 18