Jérémy Dufloo
@jeremydufloo.bsky.social
300 followers
350 following
35 posts
Postdoc in Rafael Sanjuán's lab (Universidad de Valencia) studying viral emergence experimentally | Former PhD in Olivier Schwartz's lab (Institut Pasteur) studying antibody responses against HIV-1 and SARS-CoV-2
Posts
Media
Videos
Starter Packs
Jérémy Dufloo
@jeremydufloo.bsky.social
· Aug 23
Jérémy Dufloo
@jeremydufloo.bsky.social
· Aug 18

Identification and characterization of novel bat coronaviruses in Spain
Author summary Bats carry many different viruses, some of which can infect humans. Among these, bat coronaviruses are of particular concern. To be better prepared for future pandemics, it is important...
dx.plos.org
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jun 19
Reposted by Jérémy Dufloo
The Rey Lab
@virusfusion007.bsky.social
· Jan 25

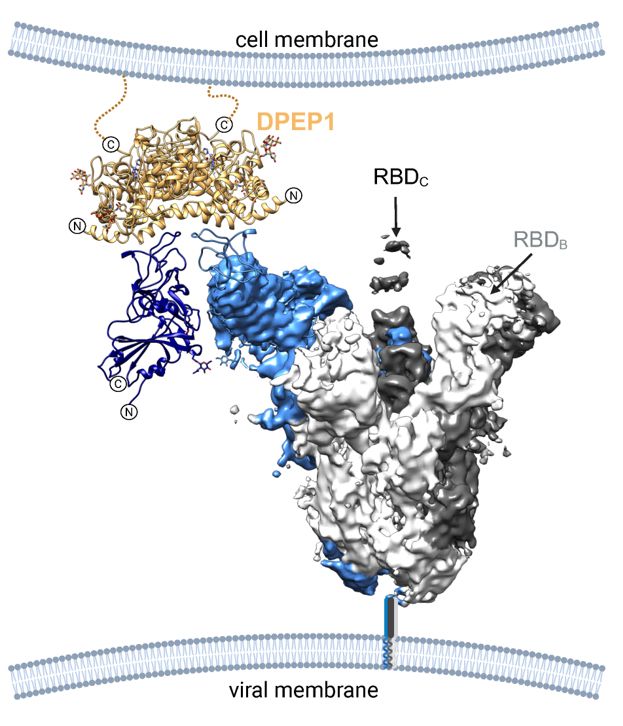
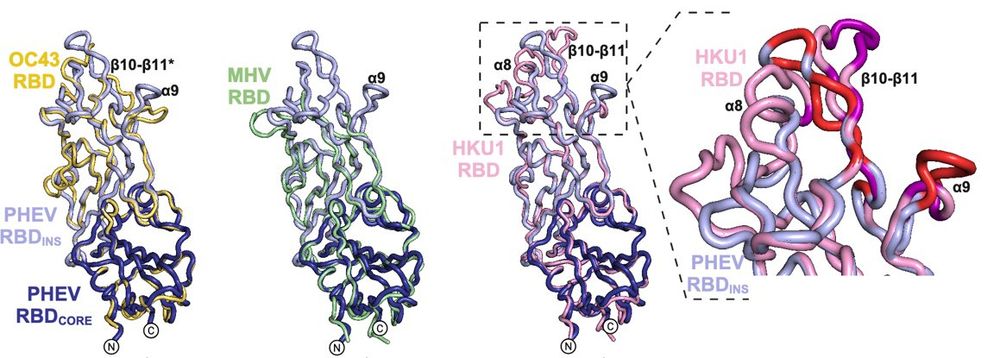
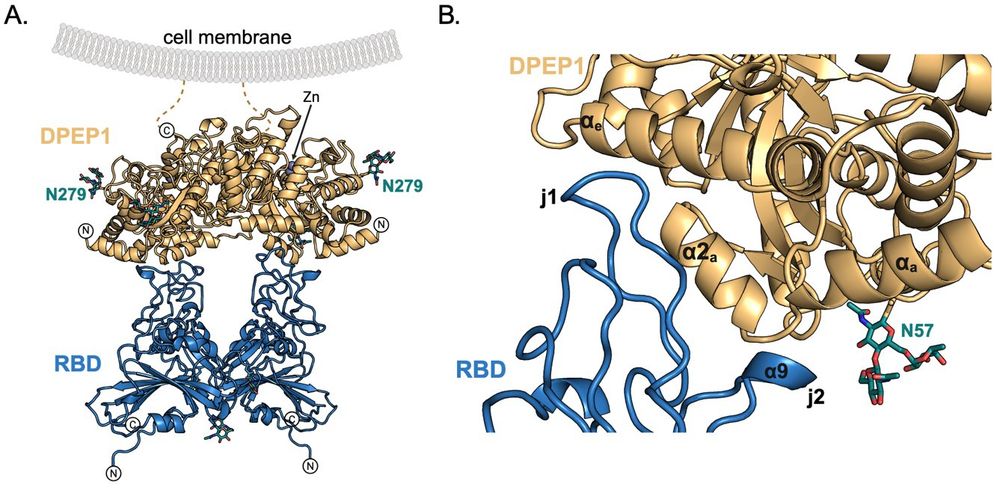
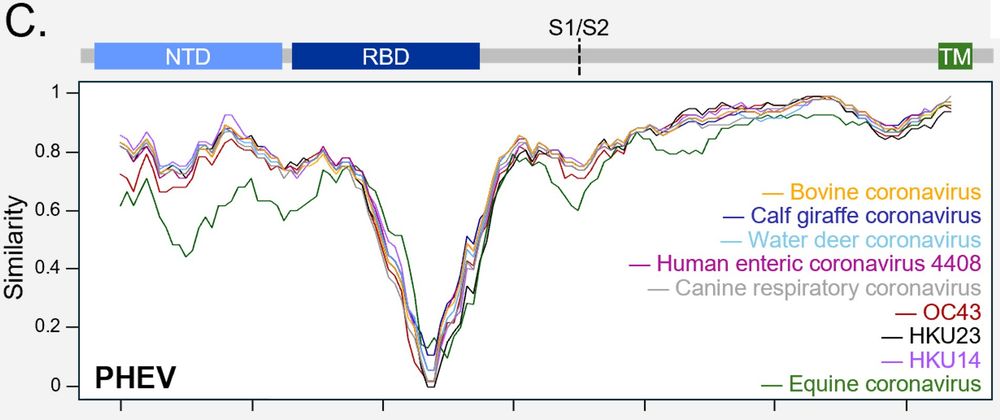
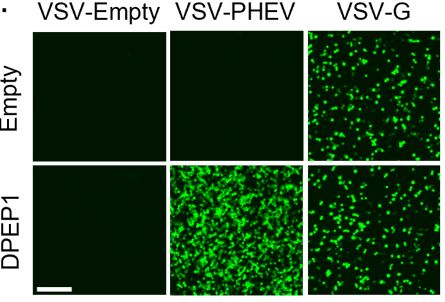
Dipeptidase 1 is a functional receptor for coronavirus PHEV
Coronaviruses of the subgenus Embecovirus include several relevant pathogens such as the human seasonal coronaviruses HKU1 and OC43, bovine coronavirus, and porcine hemagglutinating encephalomyelitis ...
www.biorxiv.org
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 23

Human cells have few barriers to prevent the entry of animal viruses
A research team from the Institute of Integrative Systems Biology (I2SysBio), a joint centre of the University of Valencia and the Spanish National Research Council (CSIC), asserts that the proteins r...
www.uv.es
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 11
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10
Jérémy Dufloo
@jeremydufloo.bsky.social
· Jan 10

Dipeptidase 1 is a functional receptor for coronavirus PHEV
Coronaviruses of the subgenus Embecovirus include several relevant pathogens such as the human seasonal coronaviruses HKU1 and OC43, bovine coronavirus, and porcine hemagglutinating encephalomyelitis ...
www.biorxiv.org