Jaime Huerta-Cepas
@jhcepas.bsky.social
150 followers
60 following
11 posts
Microbiome, evolution, comparative genomics, biodiversity and a little bit of ecology: cgmlab.org
Group Leader at CBGP-CSIC/UPM in Madrid, former scientist at EMBL Heidelberg, CRG Barcelona and CIPF Valencia.
Posts
Media
Videos
Starter Packs
Reposted by Jaime Huerta-Cepas
Reposted by Jaime Huerta-Cepas
Reposted by Jaime Huerta-Cepas
Reposted by Jaime Huerta-Cepas
DEEMteam_Orsay
@deemteam.bsky.social
· Jun 17
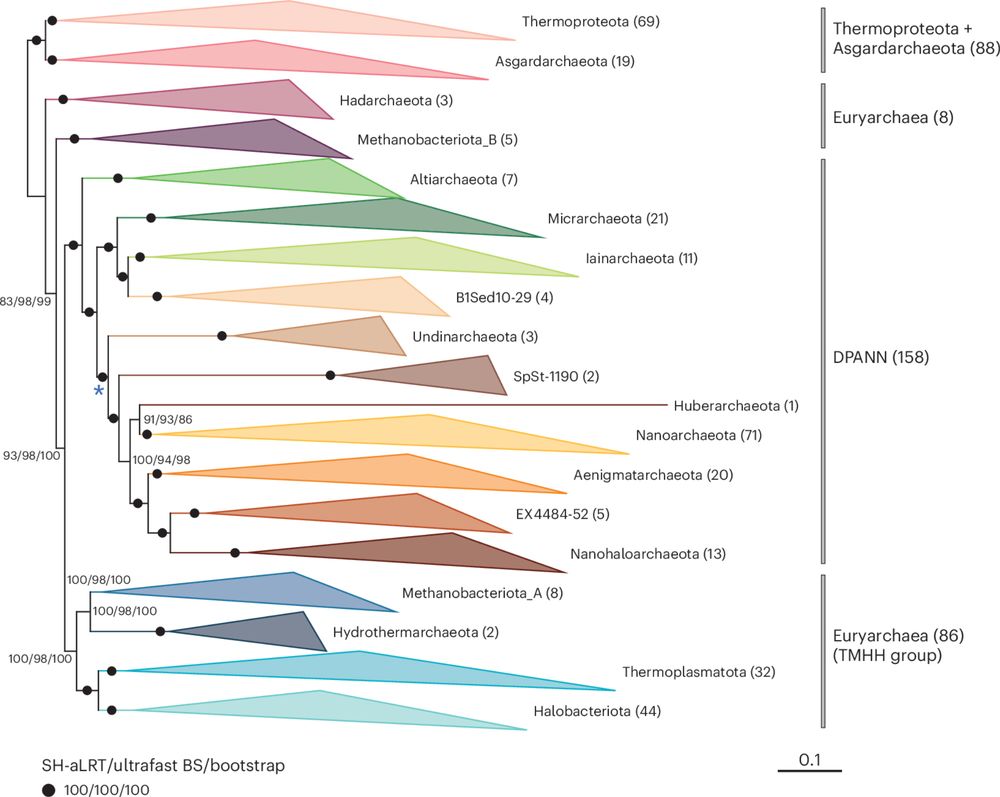
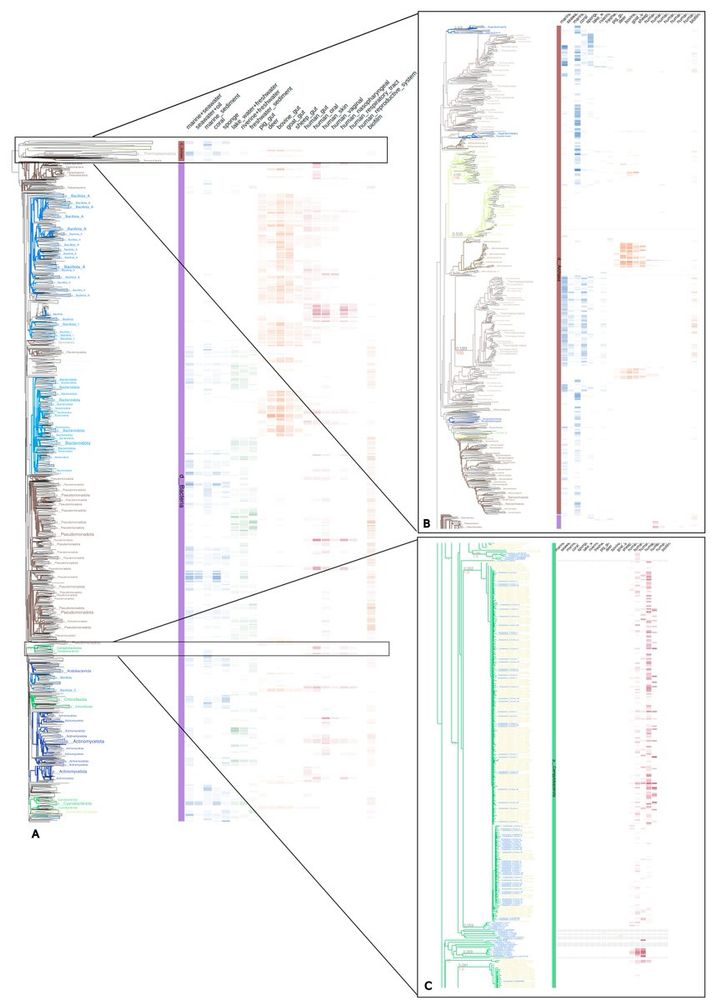
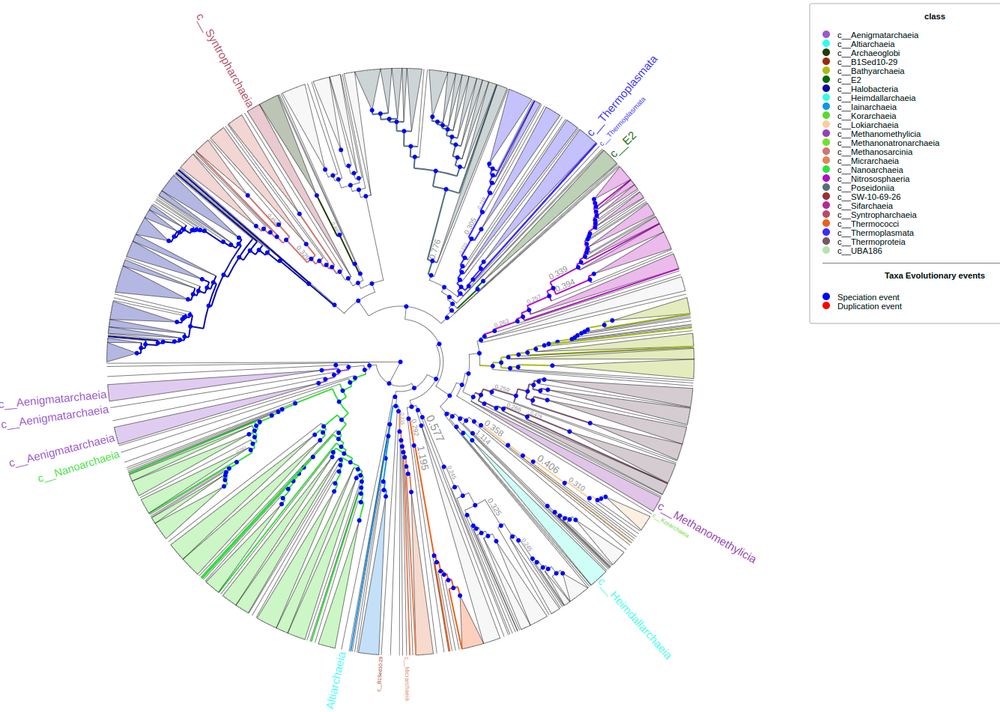
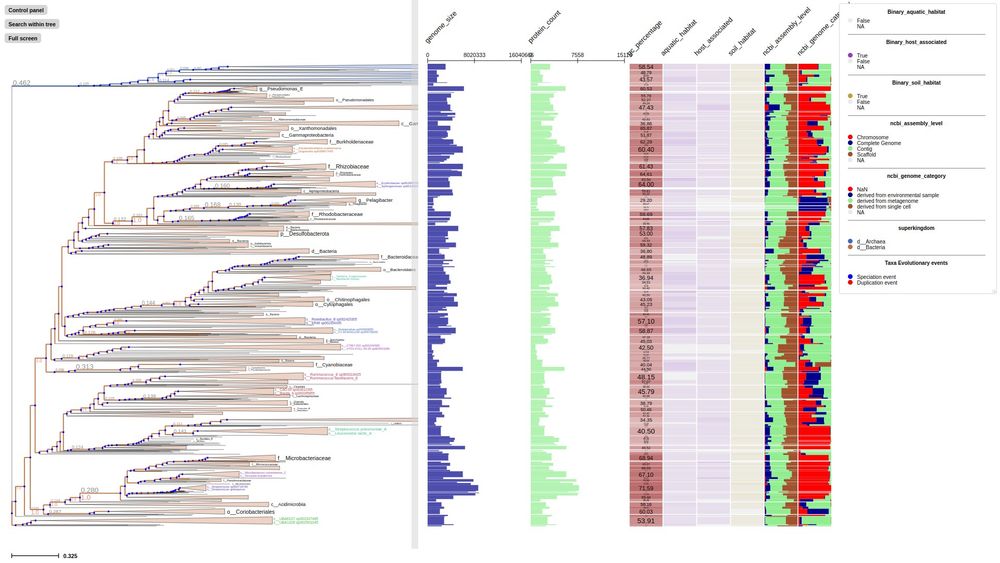
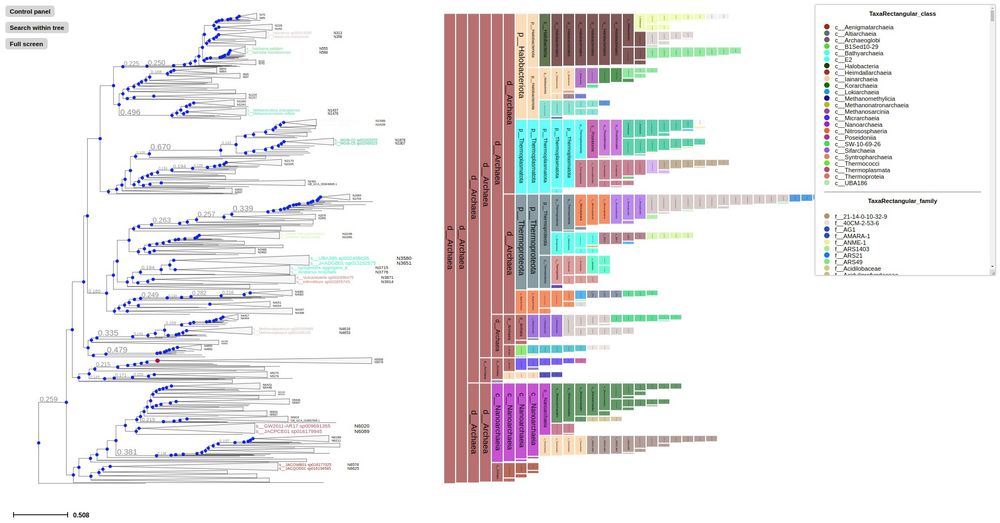
Phylogenomic analyses indicate the archaeal superphylum DPANN originated from free-living euryarchaeal-like ancestors
Nature Microbiology - Phylogenetic reconstructions with conserved protein markers from the 11 known DPANN phyla reveal their monophyletic placement within the Euryarchaeota.
rdcu.be
Reposted by Jaime Huerta-Cepas