Altogether, we could demonstrate the utility of ancient enamel proteins for phylogenetic analysis. At the example of primates, we uncovered potential limitations and pitfalls and provide recommendations on how to address them.
academic.oup.com/gbe/article/...

Altogether, we could demonstrate the utility of ancient enamel proteins for phylogenetic analysis. At the example of primates, we uncovered potential limitations and pitfalls and provide recommendations on how to address them.
academic.oup.com/gbe/article/...
We simulated ancient data by only using fragmentary sequence information and performed phylogenetic analysis on different degrees of fragmentation. A correct placement of nearly all family and genera was possible with as little as 12% of the full-length sequence data (panel c).
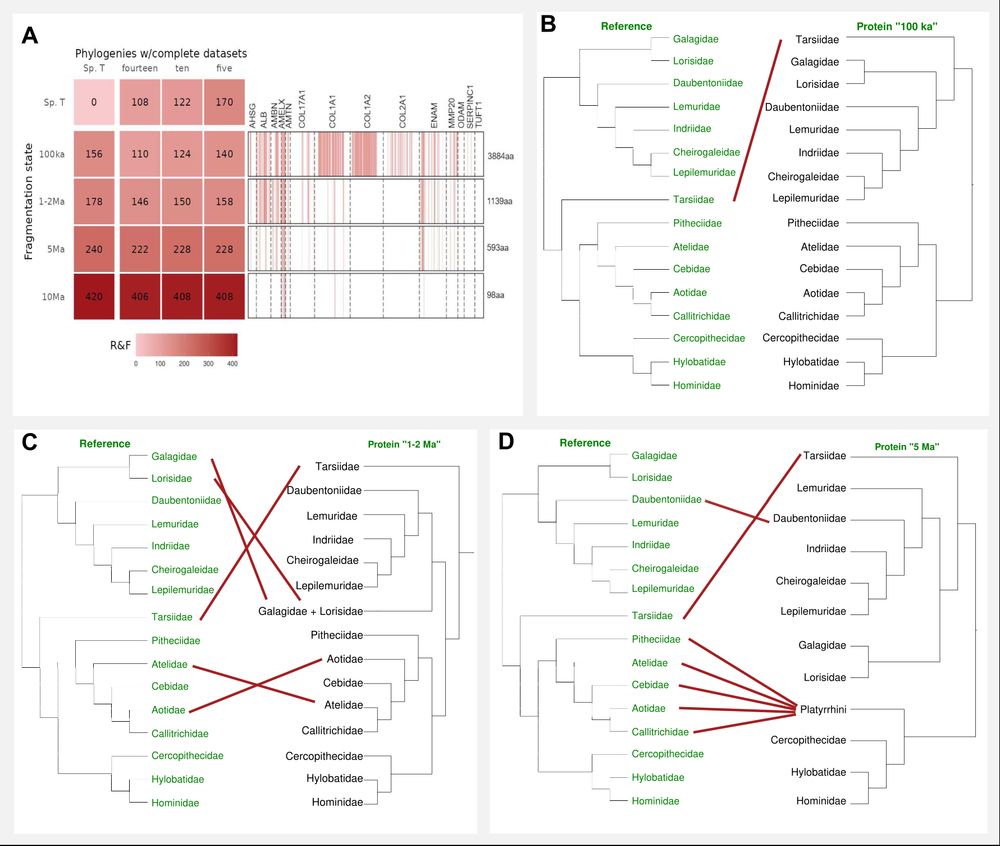
We simulated ancient data by only using fragmentary sequence information and performed phylogenetic analysis on different degrees of fragmentation. A correct placement of nearly all family and genera was possible with as little as 12% of the full-length sequence data (panel c).
One big surprise was that the deepest split in primates was affected by whether collagens belong to the proteins used in the phylogenetic analysis. The inclusion of particular collagen sequences caused a placement of tarsiers with strepsirrhines - a placement that nowadays is widely rejected.
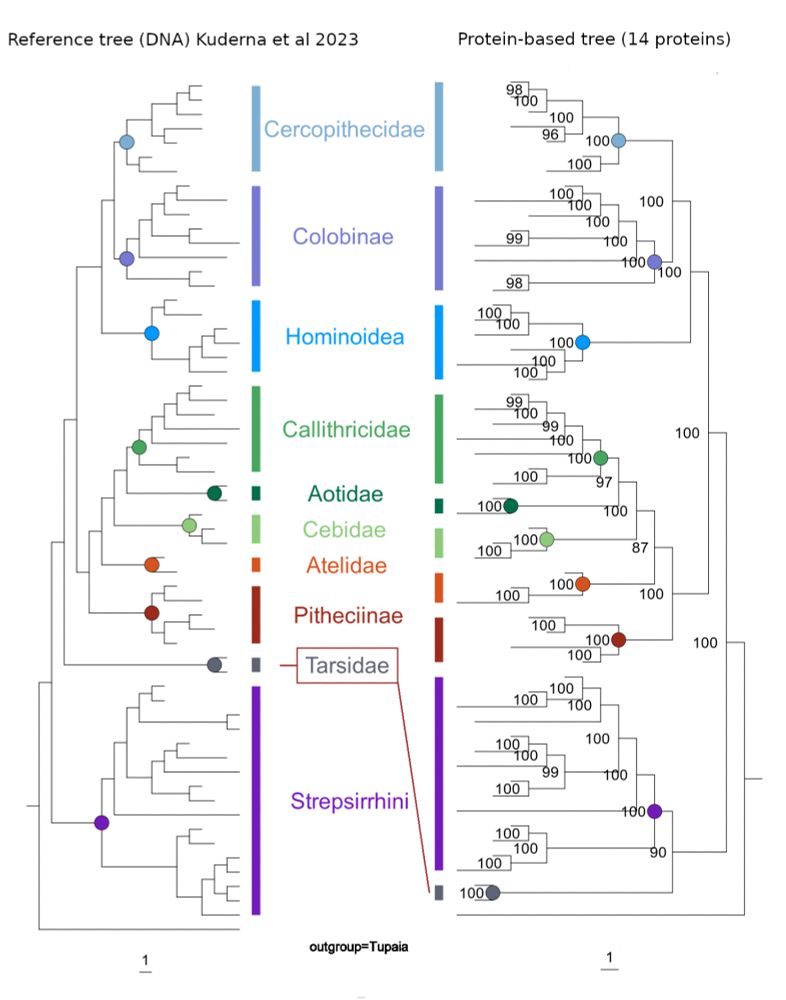
One big surprise was that the deepest split in primates was affected by whether collagens belong to the proteins used in the phylogenetic analysis. The inclusion of particular collagen sequences caused a placement of tarsiers with strepsirrhines - a placement that nowadays is widely rejected.
We translated 14 dental enamel protein sequences from 232 primate species. First, we checked how sequence conservation coincides with those peptides that are typically recovered from ancient dental enamel of different ages (purple shading).

We translated 14 dental enamel protein sequences from 232 primate species. First, we checked how sequence conservation coincides with those peptides that are typically recovered from ancient dental enamel of different ages (purple shading).
Dental enamel has a small proteome that survives into deep time in fossilized teeth. Recently, ancient enamel peptides could be isolated, sequenced, and used for phylogenetic analysis. However, it is unclear how much and which sequence information is needed for reliable phylogenetic inference…
Dental enamel has a small proteome that survives into deep time in fossilized teeth. Recently, ancient enamel peptides could be isolated, sequenced, and used for phylogenetic analysis. However, it is unclear how much and which sequence information is needed for reliable phylogenetic inference…
Dental enamel has a small proteome that survives into deep time in fossilized teeth. Recently, ancient enamel peptides could be isolated, sequenced, and used for phylogenetic analysis. However, it is unclear how much and which sequence information is needed for reliable phylogenetic inference…
Dental enamel has a small proteome that survives into deep time in fossilized teeth. Recently, ancient enamel peptides could be isolated, sequenced, and used for phylogenetic analysis. However, it is unclear how much and which sequence information is needed for reliable phylogenetic inference…
We translated 14 dental enamel protein sequences from 232 primate species. First, we checked how sequence conservation coincides with those peptides that are typically recovered from ancient dental enamel of different ages (purple shading).

We translated 14 dental enamel protein sequences from 232 primate species. First, we checked how sequence conservation coincides with those peptides that are typically recovered from ancient dental enamel of different ages (purple shading).
One big surprise was that the deepest split in primates was affected by whether collagens belong to the proteins used in the phylogenetic analysis. The inclusion of particular collagen sequences caused a placement of tarsiers with strepsirrhines - a placement that nowadays is widely rejected.

One big surprise was that the deepest split in primates was affected by whether collagens belong to the proteins used in the phylogenetic analysis. The inclusion of particular collagen sequences caused a placement of tarsiers with strepsirrhines - a placement that nowadays is widely rejected.
We simulated ancient data by only using fragmentary sequence information and performed phylogenetic analysis on different degrees of fragmentation. A correct placement of nearly all family and genera was possible with as little as 12% of the full-length sequence data (panel c).

We simulated ancient data by only using fragmentary sequence information and performed phylogenetic analysis on different degrees of fragmentation. A correct placement of nearly all family and genera was possible with as little as 12% of the full-length sequence data (panel c).
Altogether, we could demonstrate the utility of ancient enamel proteins for phylogenetic analysis. At the example of primates, we uncovered potential limitations and pitfalls and provide recommendations on how to address them.
academic.oup.com/gbe/article/...

Altogether, we could demonstrate the utility of ancient enamel proteins for phylogenetic analysis. At the example of primates, we uncovered potential limitations and pitfalls and provide recommendations on how to address them.
academic.oup.com/gbe/article/...
Altogether, we could demonstrate the utility of ancient enamel proteins for phylogenetic analysis. At the example of primates, we uncovered potential limitations and pitfalls and provide recommendations on how to address them.
academic.oup.com/gbe/article/...

Altogether, we could demonstrate the utility of ancient enamel proteins for phylogenetic analysis. At the example of primates, we uncovered potential limitations and pitfalls and provide recommendations on how to address them.
academic.oup.com/gbe/article/...
We translated 14 dental enamel protein sequences from 232 primate species. First, we checked how sequence conservation coincides with those peptides that are typically recovered from ancient dental enamel of different ages (purple shading).

We translated 14 dental enamel protein sequences from 232 primate species. First, we checked how sequence conservation coincides with those peptides that are typically recovered from ancient dental enamel of different ages (purple shading).
Dental enamel has a small proteome that survives into deep time in fossilized teeth. Recently, ancient enamel peptides could be isolated, sequenced, and used for phylogenetic analysis. However, it is unclear how much and which sequence information is needed for reliable phylogenetic inference…
Dental enamel has a small proteome that survives into deep time in fossilized teeth. Recently, ancient enamel peptides could be isolated, sequenced, and used for phylogenetic analysis. However, it is unclear how much and which sequence information is needed for reliable phylogenetic inference…

