Jan Řezáč
@jrezac.bsky.social
120 followers
60 following
18 posts
Computational chemist at @iocbprague.bsky.social
Posts
Media
Videos
Starter Packs
Reposted by Jan Řezáč
Reposted by Jan Řezáč
Jan Řezáč
@jrezac.bsky.social
· Jul 4
Jan Řezáč
@jrezac.bsky.social
· Jul 2
Jan Řezáč
@jrezac.bsky.social
· Jul 2
Jan Řezáč
@jrezac.bsky.social
· Jul 2
Jan Řezáč
@jrezac.bsky.social
· Jun 26
Jan Řezáč
@jrezac.bsky.social
· May 20

Multiscale Computational Protocols for Accurate Residue Interactions at the Flexible Insulin–Receptor Interface
The quantitative characterization of residue contributions to protein–protein binding across extensive flexible interfaces poses a significant challenge for biophysical computations. It is attributabl...
pubs.acs.org
Reposted by Jan Řezáč
Jan Řezáč
@jrezac.bsky.social
· Mar 17

Comparative Analysis of Quantum-Mechanical and standard Single-Structure Protein-Ligand Scoring Functions with MD-Based Free Energy Calculations
Single-structure scoring functions have been considered inferior to expensive ensemble free energy methods in predicting protein-ligand affinities. We are revisiting this dogma with the recently devel...
doi.org
Jan Řezáč
@jrezac.bsky.social
· Mar 10
Adam Pecina
@adampecina.bsky.social
· Mar 10
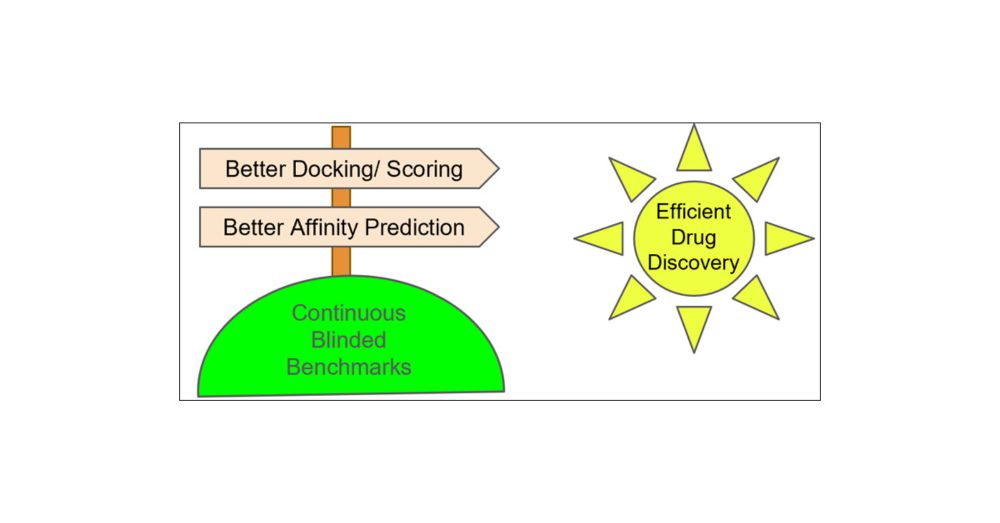
The Need for Continuing Blinded Pose- and Activity Prediction Benchmarks
Computational tools for structure-based drug design (SBDD) are widely used in drug discovery and can provide valuable insights to advance projects in an efficient and cost-effective manner. However, d...
doi.org
Jan Řezáč
@jrezac.bsky.social
· Jan 15
Jan Řezáč
@jrezac.bsky.social
· Jan 13

PM6-ML: The Synergy of Semiempirical Quantum Chemistry and Machine Learning Transformed into a Practical Computational Method
Machine learning (ML) methods offer a promising route to the construction of universal molecular potentials with high accuracy and low computational cost. It is becoming evident that integrating physi...
pubs.acs.org