JT Neal
@jtneal.bsky.social
1.1K followers
610 following
35 posts
PI @ http://neallab.org. Scientist building tools to understand genetic variation in health and disease. Trail runner & World’s Okayest Dad.
Posts
Media
Videos
Starter Packs
Pinned
JT Neal
@jtneal.bsky.social
· Jan 27
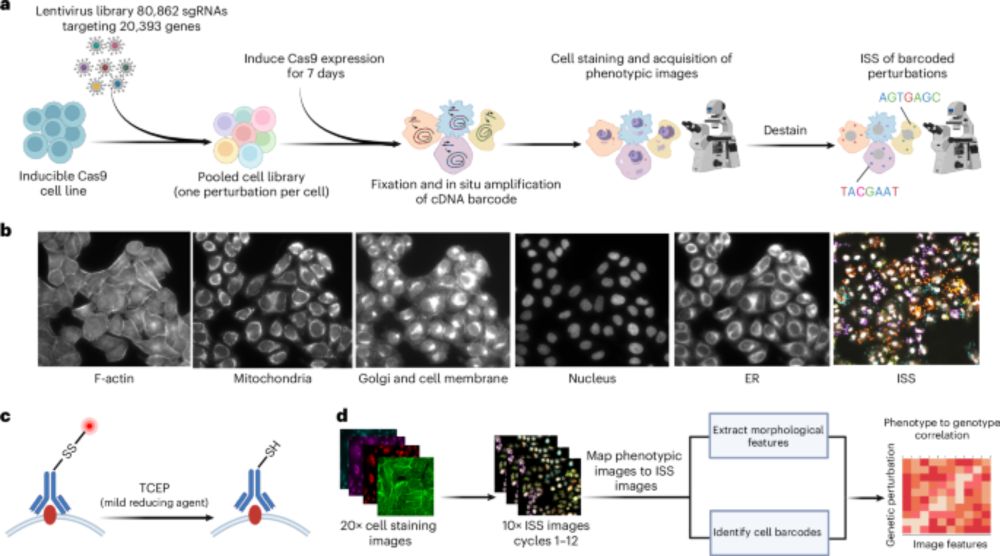
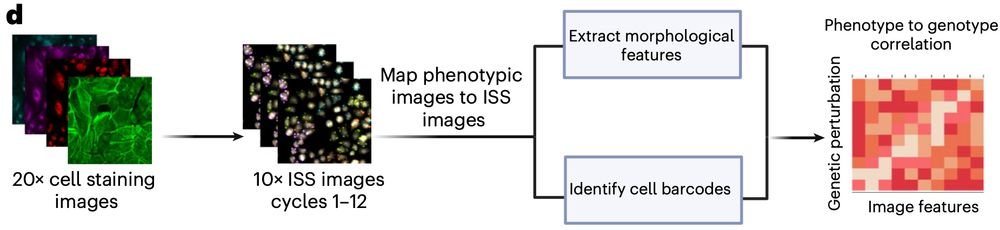
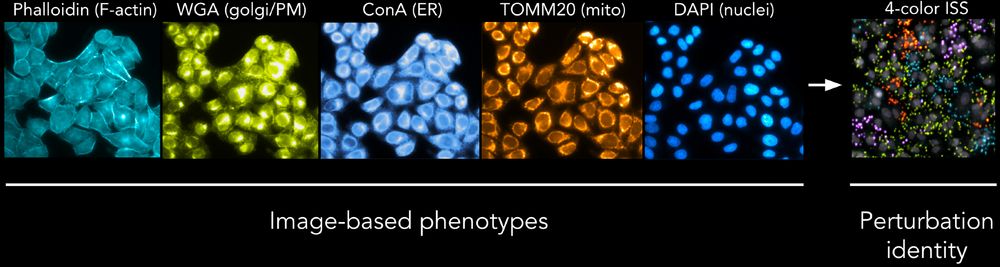
A genome-wide atlas of human cell morphology - Nature Methods
An optical pooled cell profiling platform (PERISCOPE) based on Cell Painting and optical sequencing of molecular barcodes was used to develop the first unbiased genome-wide morphology-based perturbati...
www.nature.com
Reposted by JT Neal
Reposted by JT Neal
Maitreyi Das
@daslabpombe.com
· Feb 7
JT Neal
@jtneal.bsky.social
· Jan 29
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27
JT Neal
@jtneal.bsky.social
· Jan 27

Mapping information-rich genotype-phenotype landscapes with genome-scale Perturb-seq
Unbiased, genome-scaling profiling of genetic perturbations via single-cell RNA sequencing
enables systematic assignment of function to genes and in-depth study of complex cellular
phenotypes such as ...
www.cell.com
JT Neal
@jtneal.bsky.social
· Jan 27