Jae Young Choi
@jychoi.bsky.social
260 followers
390 following
28 posts
Assistant professor of evolutionary genomics at KU. Has a toddler 🙂🙃🙂🙃🇰🇷🇨🇦🇺🇲
https://jychoilab.github.io/
Posts
Media
Videos
Starter Packs
Jae Young Choi
@jychoi.bsky.social
· Jul 15

Topsicle: a method for estimating telomere length from whole genome long-read sequencing data
Telomeres protect chromosome ends and its length varies significantly between organisms. Because telomere length variation is associated with various biomedical and eco-evolutionary phenotypes, many b...
www.biorxiv.org
Jae Young Choi
@jychoi.bsky.social
· Jul 30
Reposted by Jae Young Choi
Li Zhao
@lizhao.bsky.social
· Jul 15
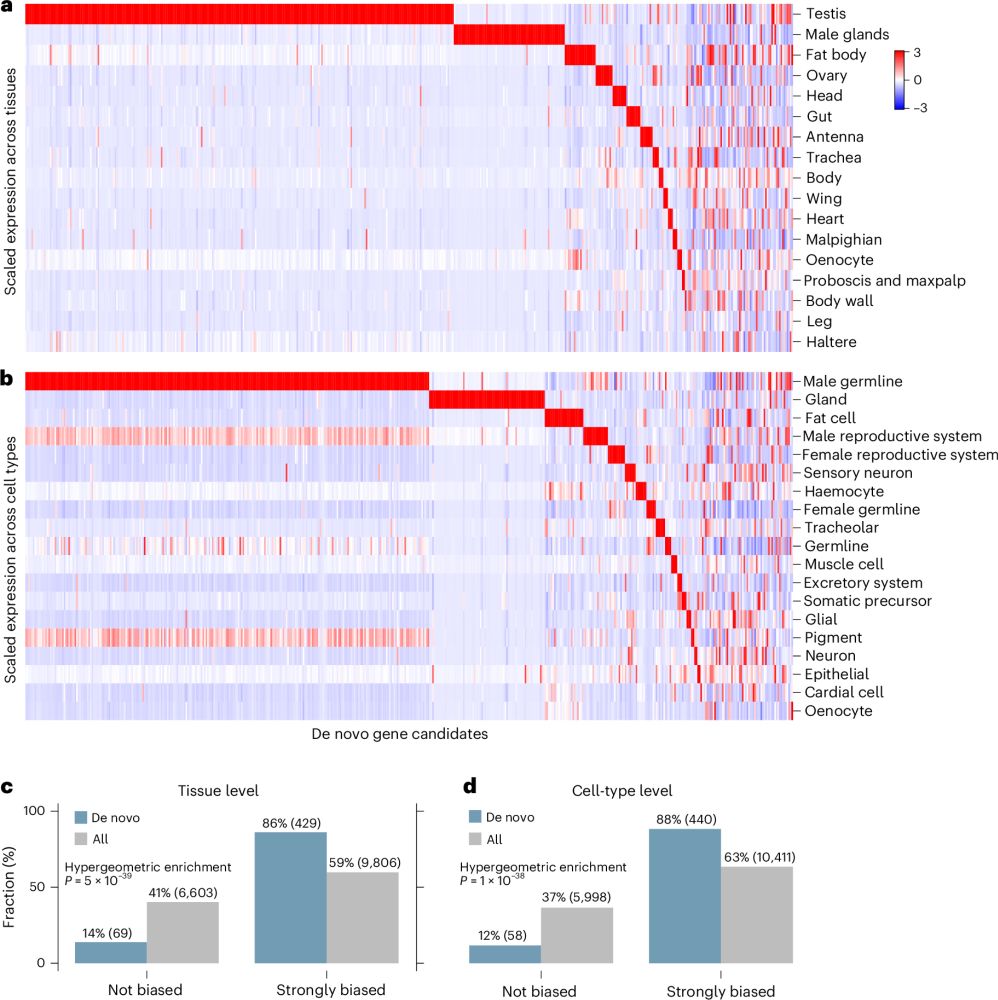
Gene regulatory networks and essential transcription factors for de novo-originated genes
Nature Ecology & Evolution - A combination of computational methods applied to single-cell RNA sequencing data and genetic experiments shows that a small number of transcription factors are...
rdcu.be
Jae Young Choi
@jychoi.bsky.social
· Jul 15

GitHub - jaeyoungchoilab/Topsicle: Topsicle utilizes abundance of telomere pattern k-mers to estimate telomere length in long read.
Topsicle utilizes abundance of telomere pattern k-mers to estimate telomere length in long read. - jaeyoungchoilab/Topsicle
github.com
Jae Young Choi
@jychoi.bsky.social
· Jul 15
Jae Young Choi
@jychoi.bsky.social
· Jul 15

Topsicle: a method for estimating telomere length from whole genome long-read sequencing data
Telomeres protect chromosome ends and its length varies significantly between organisms. Because telomere length variation is associated with various biomedical and eco-evolutionary phenotypes, many b...
www.biorxiv.org
Jae Young Choi
@jychoi.bsky.social
· Jun 28
Jae Young Choi
@jychoi.bsky.social
· Jun 28

GitHub - jaeyoungchoilab/Topsicle: Topsicle utilizes abundance of telomere pattern k-mers to estimate telomere length in long read.
Topsicle utilizes abundance of telomere pattern k-mers to estimate telomere length in long read. - jaeyoungchoilab/Topsicle
github.com
Jae Young Choi
@jychoi.bsky.social
· Jun 17
Jae Young Choi
@jychoi.bsky.social
· Jun 17

Monkeyflower (Mimulus) uncovers the evolutionary basis of the eukaryote telomere sequence variation
Author summary One of the most fascinating phenomena in evolutionary biology is the rapid evolution of molecular complexes with conserved functions across the tree of life. Studying these complexes ha...
journals.plos.org
Reposted by Jae Young Choi

















