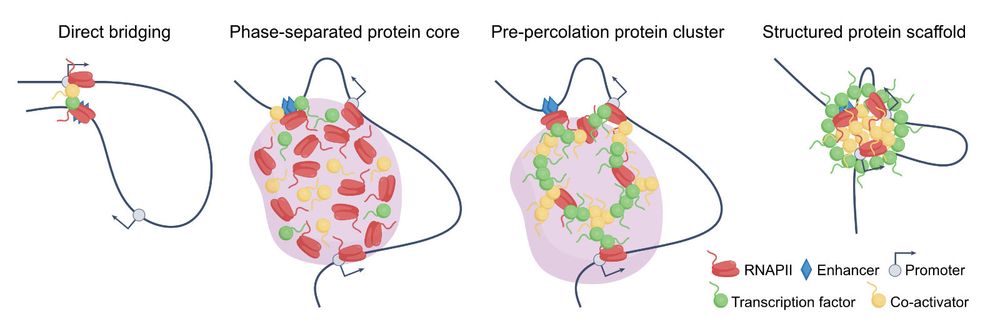
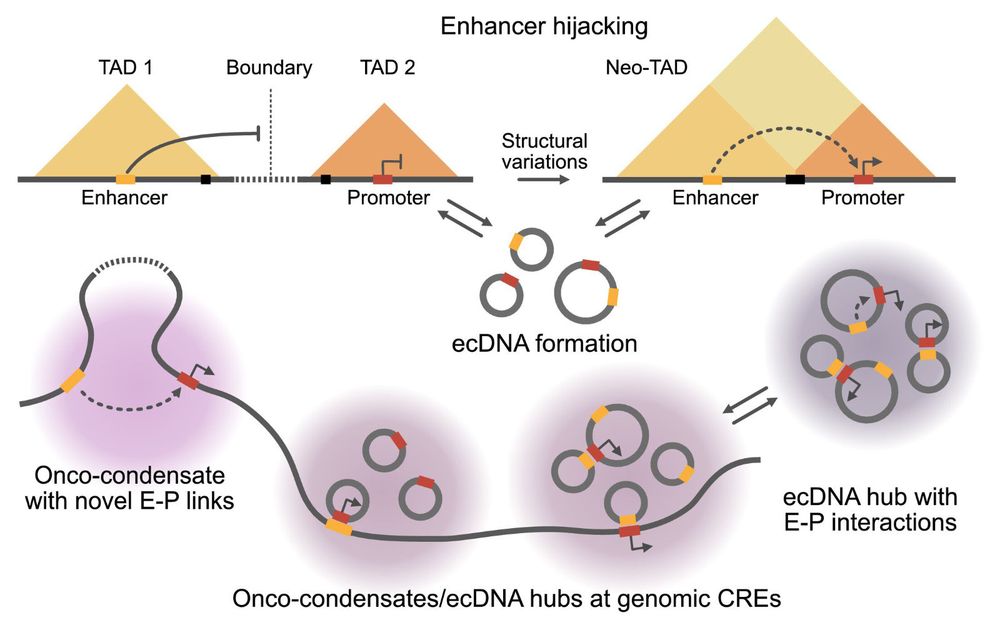
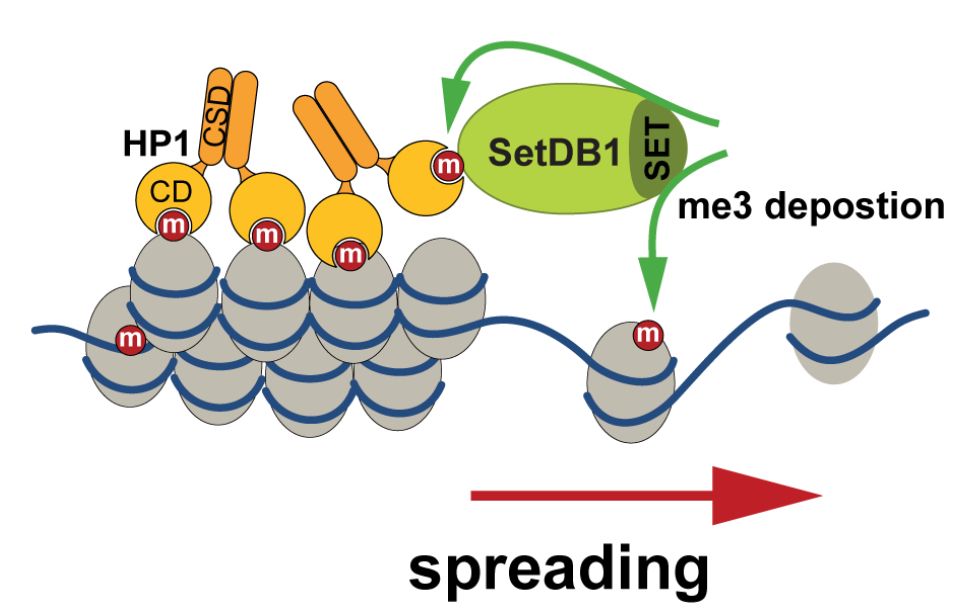
Karsten Rippe
@karsten-rippe.bsky.social
460 followers
210 following
27 posts
https://malone.bioquant.uni-heidelberg.de
Posts
Media
Videos
Starter Packs
Karsten Rippe
@karsten-rippe.bsky.social
· Aug 21
Current practices in the study of biomolecular condensates: a community comment - Nature Communications
The realization that the cell is abundantly compartmentalized into biomolecular condensates has opened new opportunities for understanding the physics and chemistry underlying many cellular processes1...
doi.org
Karsten Rippe
@karsten-rippe.bsky.social
· Aug 13
Karsten Rippe
@karsten-rippe.bsky.social
· Jun 20
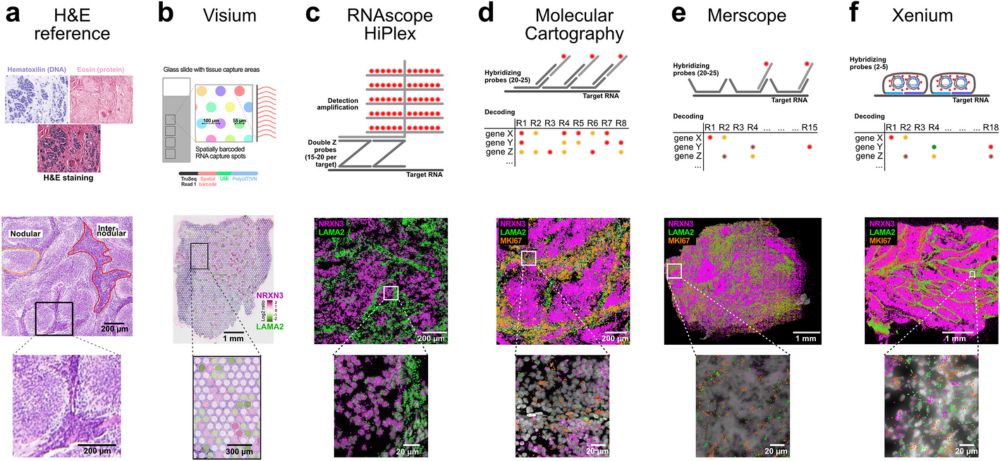
Comparison of spatial transcriptomics technologies using tumor cryosections - Genome Biology
Background Spatial transcriptomics technologies are revolutionizing our understanding of intra-tumor heterogeneity and the tumor microenvironment by revealing single-cell molecular profiles within the...
doi.org
Karsten Rippe
@karsten-rippe.bsky.social
· Jan 22

Fakten sind das Rückgrat unserer Demokratie - Online-Petition
Liebe Mitbürgerinnen und Mitbürger, als Wissenschaftlerinnen und Wissenschaftler arbeiten wir täglich daran, die Grundlagen für fundierte Entscheidungen und Innovationen zu schaffen – basierend auf üb...
www.openpetition.de
Karsten Rippe
@karsten-rippe.bsky.social
· Dec 13
Karsten Rippe
@karsten-rippe.bsky.social
· Dec 13
Karsten Rippe
@karsten-rippe.bsky.social
· Dec 13
Karsten Rippe
@karsten-rippe.bsky.social
· Dec 13
Karsten Rippe
@karsten-rippe.bsky.social
· Dec 13
Reposted by Karsten Rippe