Posts
Media
Videos
Starter Packs
Kusterlab
@kusterlab.bsky.social
· Aug 25

Enhanced photo-crosslinking in living cells with high-intensity longwave ultraviolet light
The activation of chemical reactions in living cells using ultraviolet (UV) light enables the interrogation of biomolecules in their native environment with photoreactive probes or crosslinking reagen...
www.biorxiv.org
Kusterlab
@kusterlab.bsky.social
· Jul 31
Kusterlab
@kusterlab.bsky.social
· Jul 31
Kusterlab
@kusterlab.bsky.social
· May 22
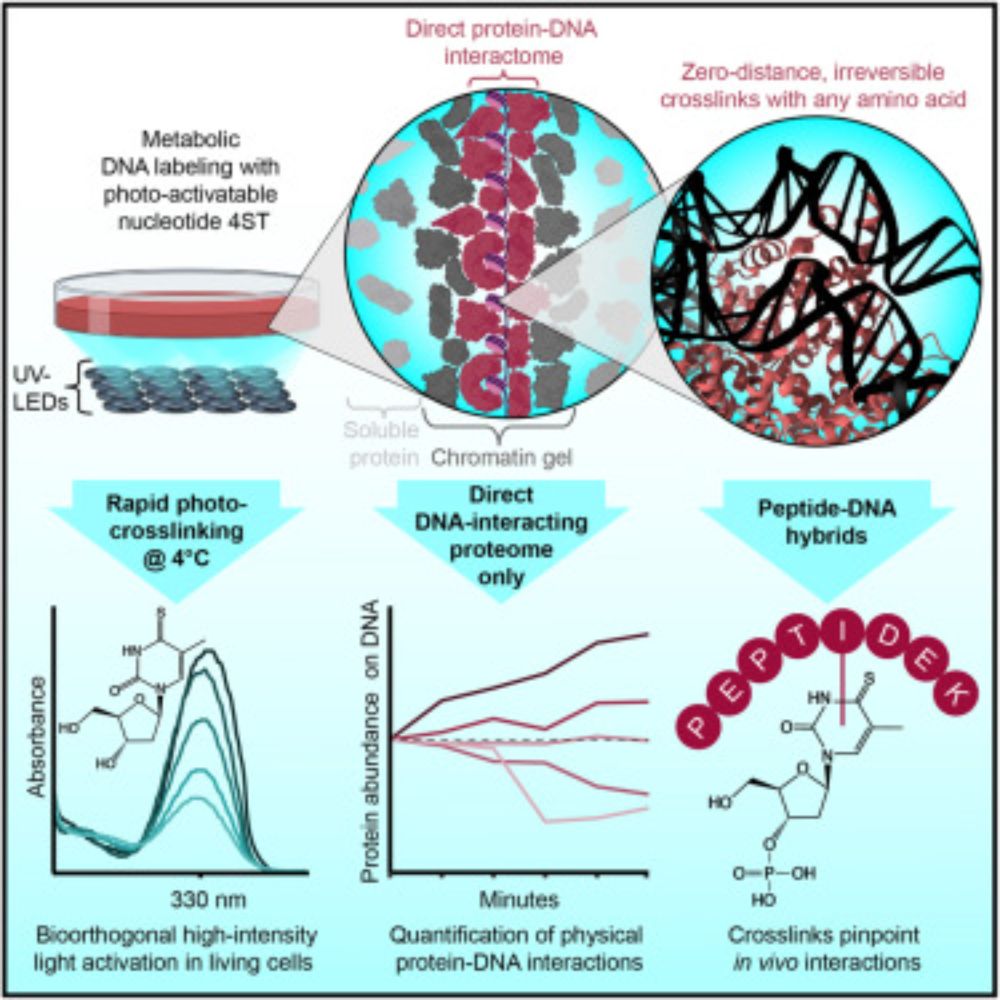
The human proteome with direct physical access to DNA
Zero-distance photo-crosslinking reveals direct protein-DNA interactions in living
cells, enabling quantitative analysis of the DNA-interacting proteome on a timescale
of minutes with single-amino-aci...
www.cell.com
Kusterlab
@kusterlab.bsky.social
· May 9
Kusterlab
@kusterlab.bsky.social
· May 9

sev on X: "Paper alert! We report the first inhibitors for nucleoside kinases NME1-4 and for the nucleotide-binding protein HINT1. On top, we provide probes for MBLAC2, an off-target of every second (!) hydroxamic acid-based HDAC inhibitor! https://t.co/uuRCO78xFz 👇🧵" / X
Paper alert! We report the first inhibitors for nucleoside kinases NME1-4 and for the nucleotide-binding protein HINT1. On top, we provide probes for MBLAC2, an off-target of every second (!) hydroxamic acid-based HDAC inhibitor! https://t.co/uuRCO78xFz 👇🧵
x.com
Kusterlab
@kusterlab.bsky.social
· Apr 22
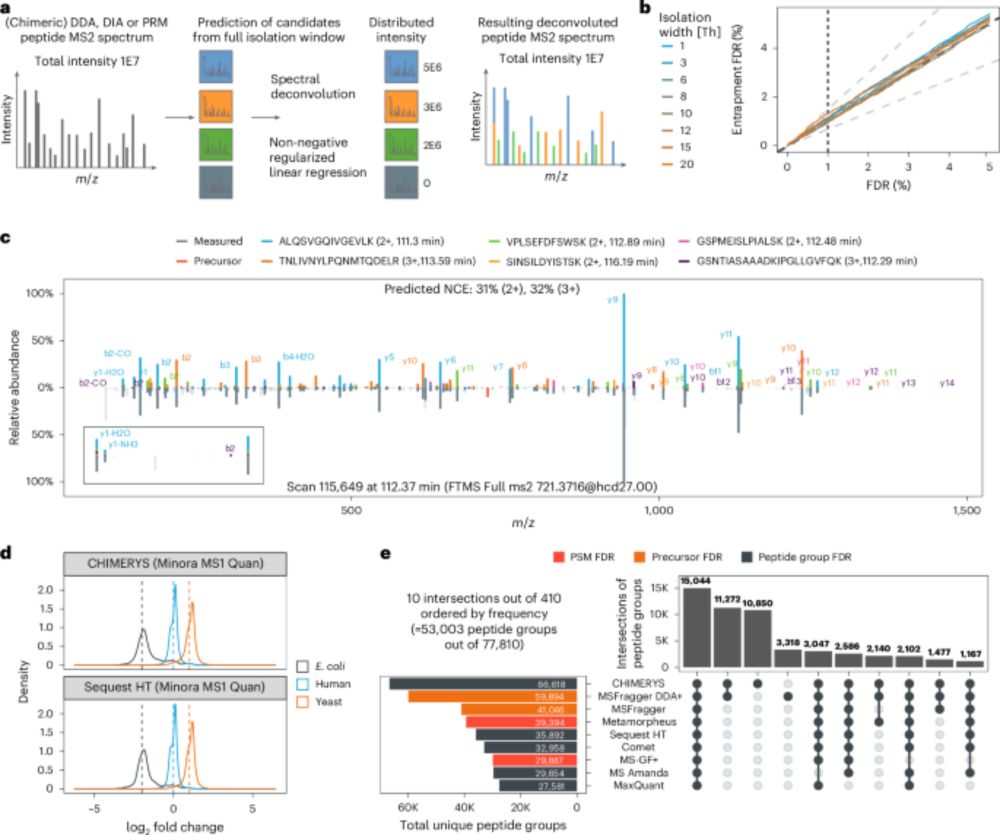
Unifying the analysis of bottom-up proteomics data with CHIMERYS - Nature Methods
CHIMERYS is a spectrum-centric and data acquisition method-agnostic algorithm for the analysis of MS2 spectra. It is capable of deconvoluting any MS2 spectrum, regardless of whether it was acquired by...
doi.org
Reposted by Kusterlab
Kusterlab
@kusterlab.bsky.social
· Mar 4

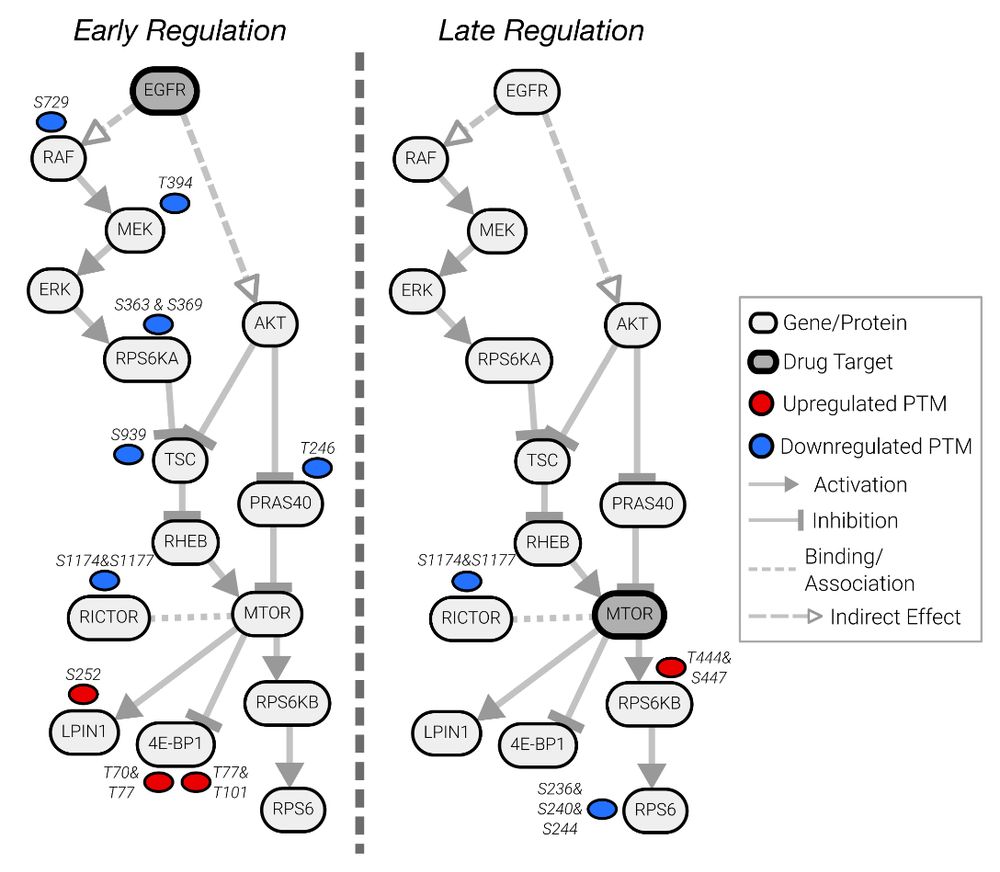
Gemcitabine and ATR inhibitors synergize to kill PDAC cells by blocking DNA damage response | Molecular Systems Biology
imageimagePhosphoproteomics unveils the mode of action of clinical ATR inhibitors and explains
their synergy with Gemcitabine in pancreatic cancer cells. Viability screening of 146 targeted drugs ide...
doi.org
Kusterlab
@kusterlab.bsky.social
· Mar 4
Kusterlab
@kusterlab.bsky.social
· Mar 4
Kusterlab
@kusterlab.bsky.social
· Mar 4

Gemcitabine and ATR inhibitors synergize to kill PDAC cells by blocking DNA damage response | Molecular Systems Biology
imageimagePhosphoproteomics unveils the mode of action of clinical ATR inhibitors and explains
their synergy with Gemcitabine in pancreatic cancer cells. Viability screening of 146 targeted drugs ide...
doi.org
Kusterlab
@kusterlab.bsky.social
· Feb 4