Luke Zappia
@lazappi.bsky.social
1K followers
320 following
45 posts
Bioinformatician, data scientist, software developer
Also @_lazappi_ and @[email protected]
Posts
Media
Videos
Starter Packs
Reposted by Luke Zappia
Reposted by Luke Zappia
Reposted by Luke Zappia
Louise Deconinck
@louisedck.bsky.social
· Aug 25
Luke Zappia
@lazappi.bsky.social
· Aug 26
Reposted by Luke Zappia
Reposted by Luke Zappia
Reposted by Luke Zappia
Reposted by Luke Zappia
Reposted by Luke Zappia
Reposted by Luke Zappia
Reposted by Luke Zappia
Yvan Saeys
@yvansaeys.bsky.social
· May 1
Reposted by Luke Zappia
Luke Zappia
@lazappi.bsky.social
· Apr 16
Luke Zappia
@lazappi.bsky.social
· Mar 20
Luke Zappia
@lazappi.bsky.social
· Mar 19
Luke Zappia
@lazappi.bsky.social
· Mar 19
Reposted by Luke Zappia
Luke Zappia
@lazappi.bsky.social
· Mar 19
Luke Zappia
@lazappi.bsky.social
· Mar 19
Reposted by Luke Zappia
Luke Zappia
@lazappi.bsky.social
· Mar 18
Luke Zappia
@lazappi.bsky.social
· Mar 18
Luke Zappia
@lazappi.bsky.social
· Mar 18

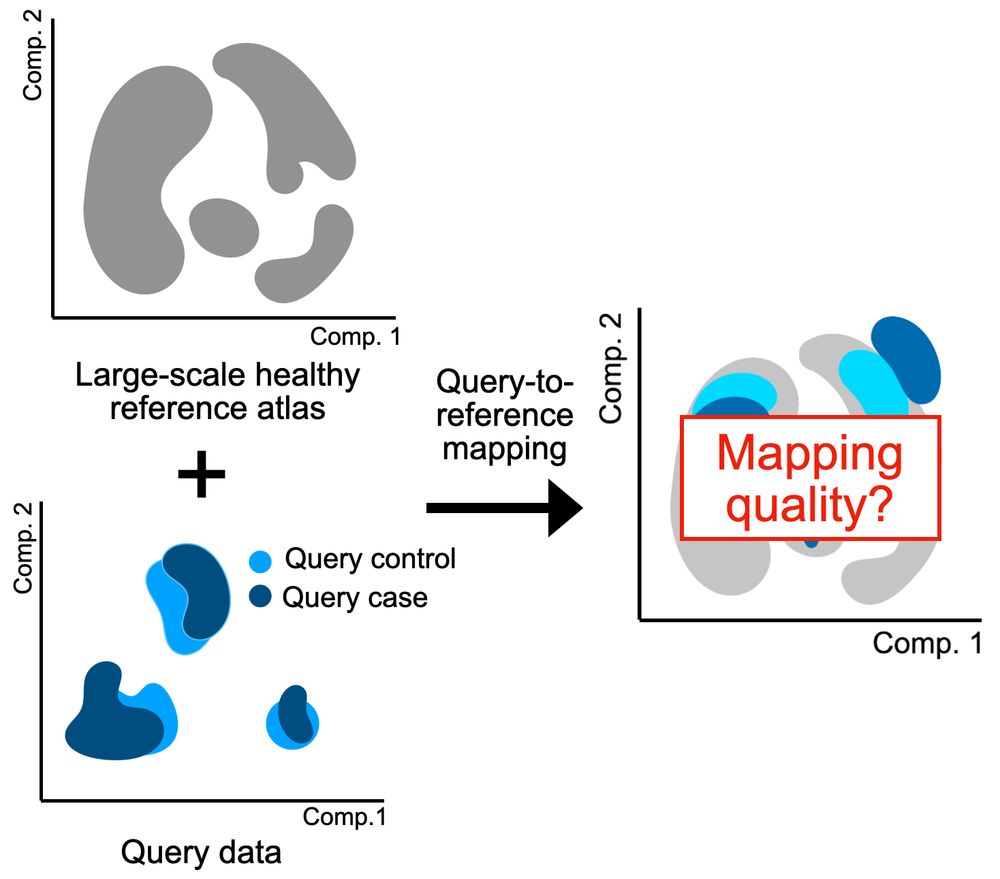
GitHub - theislab/atlas-feature-selection-benchmark: Code for "Feature selection methods affect the performance of scRNA-seq data integration and querying"
Code for "Feature selection methods affect the performance of scRNA-seq data integration and querying" - theislab/atlas-feature-selection-benchmark
github.com