Levi Waldron
@leviwaldron1.bsky.social
220 followers
13 following
22 posts
Professor of Biostatistics at CUNY SPH, rstat / Bioconductor enthusiast, cancer genomics/metagenomics, proud HPV OSCC cancer survivor.
Posts
Media
Videos
Starter Packs
Reposted by Levi Waldron
Segata Lab
@cibiocm.bsky.social
· May 16
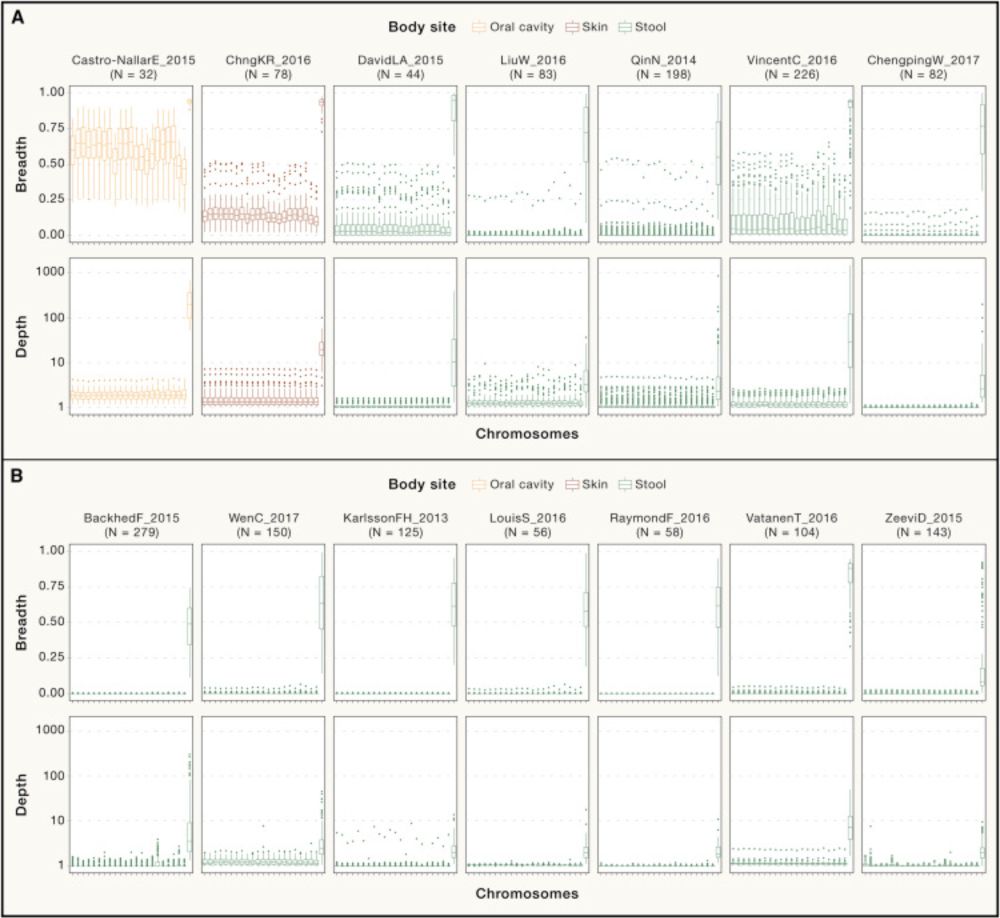
Human mitochondrial DNA in public metagenomes: Opportunity or privacy threat?
Human DNA is unavoidably present in metagenomic analyses of human microbiomes. While
current protocols remove human DNA before submission to public repositories, mitochondrial
DNA (mtDNA) has been ove...
www.cell.com
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19

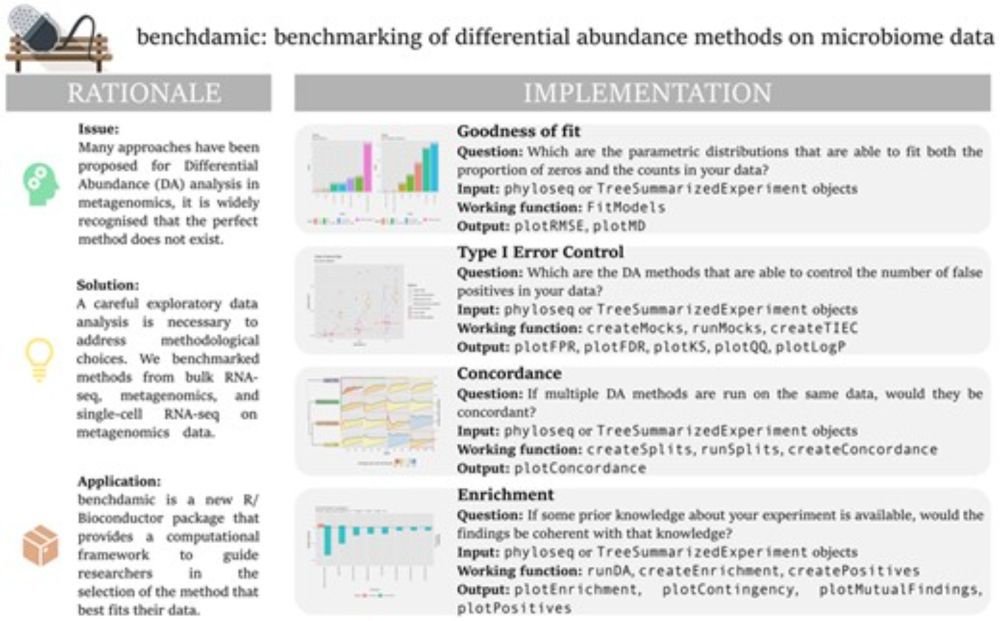
Assessment of statistical methods from single cell, bulk RNA-seq, and metagenomics applied to microbiome data - Genome Biology
Background The correct identification of differentially abundant microbial taxa between experimental conditions is a methodological and computational challenge. Recent work has produced methods to dea...
genomebiology.biomedcentral.com
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19

GitHub - waldronlab/MicrobiomeBenchmarkDataAnalyses: Analyses using the datasets provided by the MicrobiomeBenchmarkData package.
Analyses using the datasets provided by the MicrobiomeBenchmarkData package. - waldronlab/MicrobiomeBenchmarkDataAnalyses
github.com
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19

Commonly used compositional data analysis implementations are not advantageous in microbial differential abundance analyses benchmarked against biological ground truth
Previous benchmarking of differential abundance (DA) analysis methods in microbiome studies have employed synthetic data, simulations, and "real data" examples, but to the best of our knowledge, none ...
www.biorxiv.org
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19

Más-o-menos: a simple sign averaging method for discrimination in genomic data analysis
Abstract. Motivation : The successful translation of genomic signatures into clinical settings relies on good discrimination between patient subgroups. Man
academic.oup.com
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19
Levi Waldron
@leviwaldron1.bsky.social
· Feb 19

Commonly used compositional data analysis implementations are not advantageous in microbial differential abundance analyses benchmarked against biological ground truth
Previous benchmarking of differential abundance (DA) analysis methods in microbiome studies have employed synthetic data, simulations, and "real data" examples, but to the best of our knowledge, none ...
www.biorxiv.org
Reposted by Levi Waldron
Levi Waldron
@leviwaldron1.bsky.social
· Dec 21
Reposted by Levi Waldron
Reposted by Levi Waldron
Segata Lab
@cibiocm.bsky.social
· Nov 15
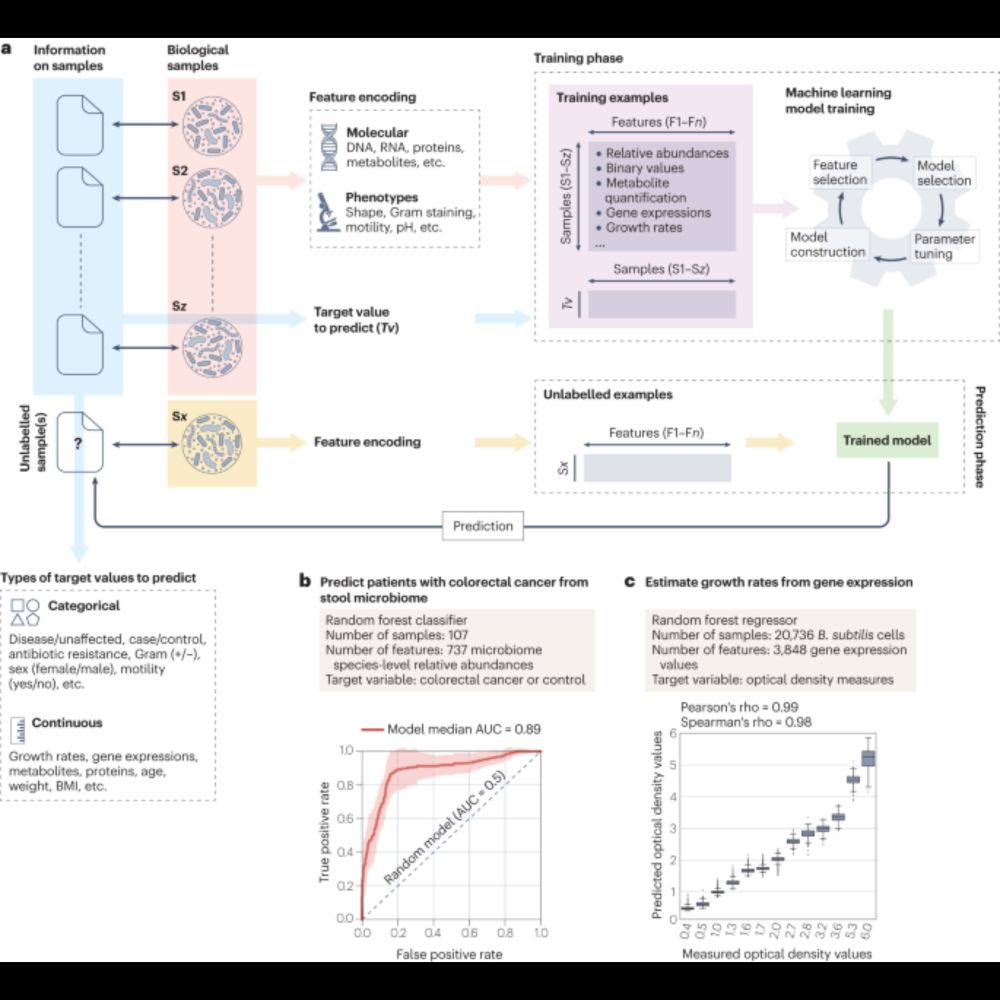
Machine learning for microbiologists - Nature Reviews Microbiology
In this Review, Segata, Waldron and colleagues discuss important key concepts of machine learning that are relevant to microbiologists and provide them with a set of tools essential to apply machine l...
www.nature.com