Loïs Rancilhac
@loisrancilhac.bsky.social
58 followers
84 following
24 posts
Evolutionary biologist: speciation, adaptation, genomics, phylogeography of birds and lizards. Keen naturalist (birder, herper, entomologist, botanist and more), climber, cyclist, hiker.
Currently in Madrid, formerly France, Germany, Sweden & Cyprus.
Posts
Media
Videos
Starter Packs
Pinned
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 26
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 14

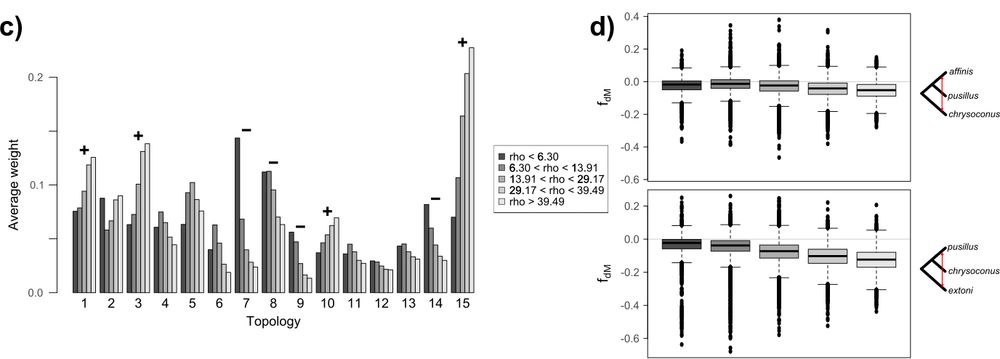
Introgression across narrow contact zones shapes the genomic landscape of phylogenetic variation in an African bird clade
Abstract. Genomic analyses of hybrid zones provide excellent opportunities to investigate the consequences of introgression in nature. In combination with
academic.oup.com
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 14
Loïs Rancilhac
@loisrancilhac.bsky.social
· May 14