Luca Pinello
@lucapinello.bsky.social
1.7K followers
830 following
29 posts
Computational methods for epigenetic, CRISPR genome editing and single-cell genomics. Associate Professor at MGH / Harvard Medical School. http://pinellolab.org
Posts
Media
Videos
Starter Packs
Reposted by Luca Pinello
Luca Pinello
@lucapinello.bsky.social
· Jul 25
Reposted by Luca Pinello
Reposted by Luca Pinello
Doug Fowler
@dougfowler.bsky.social
· Jul 21
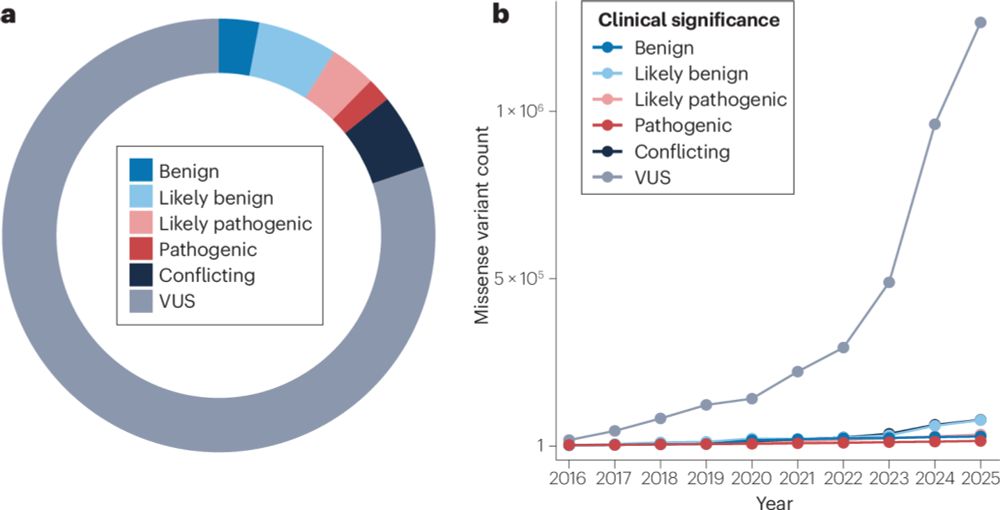
Multiplexed assays of variant effect for clinical variant interpretation
Nature Reviews Genetics - Multiplexed assays of variant effect (MAVEs) are highly scalable experimental approaches used to generate functional data for genetic variants. In this Review, McEwen et...
www.nature.com
Luca Pinello
@lucapinello.bsky.social
· Jun 8
Luca Pinello
@lucapinello.bsky.social
· Jun 8
Luca Pinello
@lucapinello.bsky.social
· Jun 8
Luca Pinello
@lucapinello.bsky.social
· Jun 8
Luca Pinello
@lucapinello.bsky.social
· Jun 8
Luca Pinello
@lucapinello.bsky.social
· Jun 8
Reposted by Luca Pinello
Reposted by Luca Pinello
Caleb Lareau
@caleblareau.bsky.social
· Jan 8
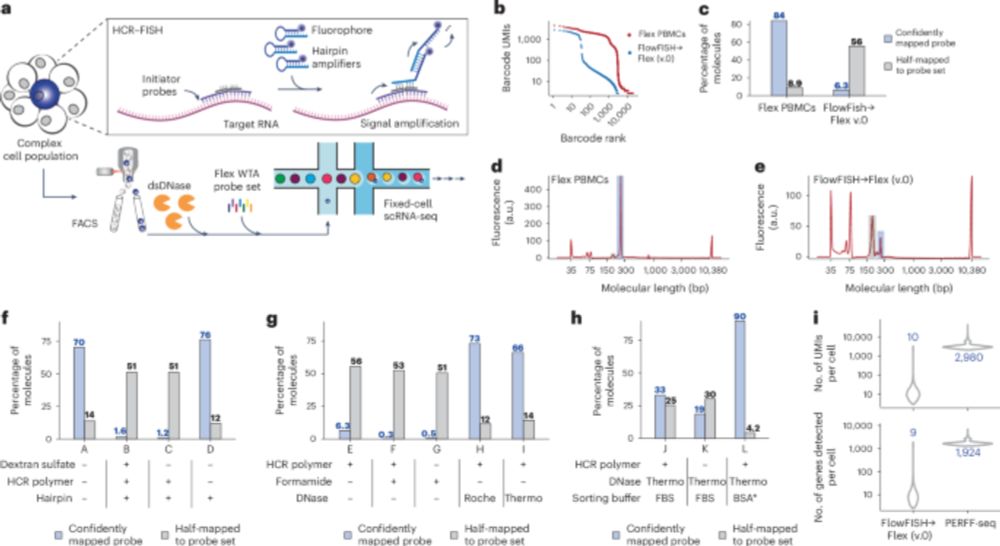
Transcript-specific enrichment enables profiling of rare cell states via single-cell RNA sequencing - Nature Genetics
Programmable Enrichment via RNA FlowFISH by sequencing (PERFF-seq) isolates rare cells based on RNA marker transcripts for single-cell RNA sequencing profiling of complex tissues, with applicability t...
www.nature.com
Luca Pinello
@lucapinello.bsky.social
· Dec 23
Luca Pinello
@lucapinello.bsky.social
· Dec 23
Luca Pinello
@lucapinello.bsky.social
· Dec 23
Luca Pinello
@lucapinello.bsky.social
· Dec 23
Luca Pinello
@lucapinello.bsky.social
· Dec 23
Luca Pinello
@lucapinello.bsky.social
· Dec 23