📍 @ml4science.bsky.social, Tübingen, Germany


We congratulate Dr Julius Vetter (@vetterj.bsky.social) and Dr Guy Moss (@gmoss13.bsky.social)! Here seen celebrating with the lab 🎳. 1/3

We congratulate Dr Julius Vetter (@vetterj.bsky.social) and Dr Guy Moss (@gmoss13.bsky.social)! Here seen celebrating with the lab 🎳. 1/3






Michael worked on "Machine Learning for Inference in Biophysical Neuroscience Simulations", focusing on simulation-based inference and differentiable simulation.
We wish him all the best for the next chapter! 👏🎓



Michael worked on "Machine Learning for Inference in Biophysical Neuroscience Simulations", focusing on simulation-based inference and differentiable simulation.
We wish him all the best for the next chapter! 👏🎓

🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
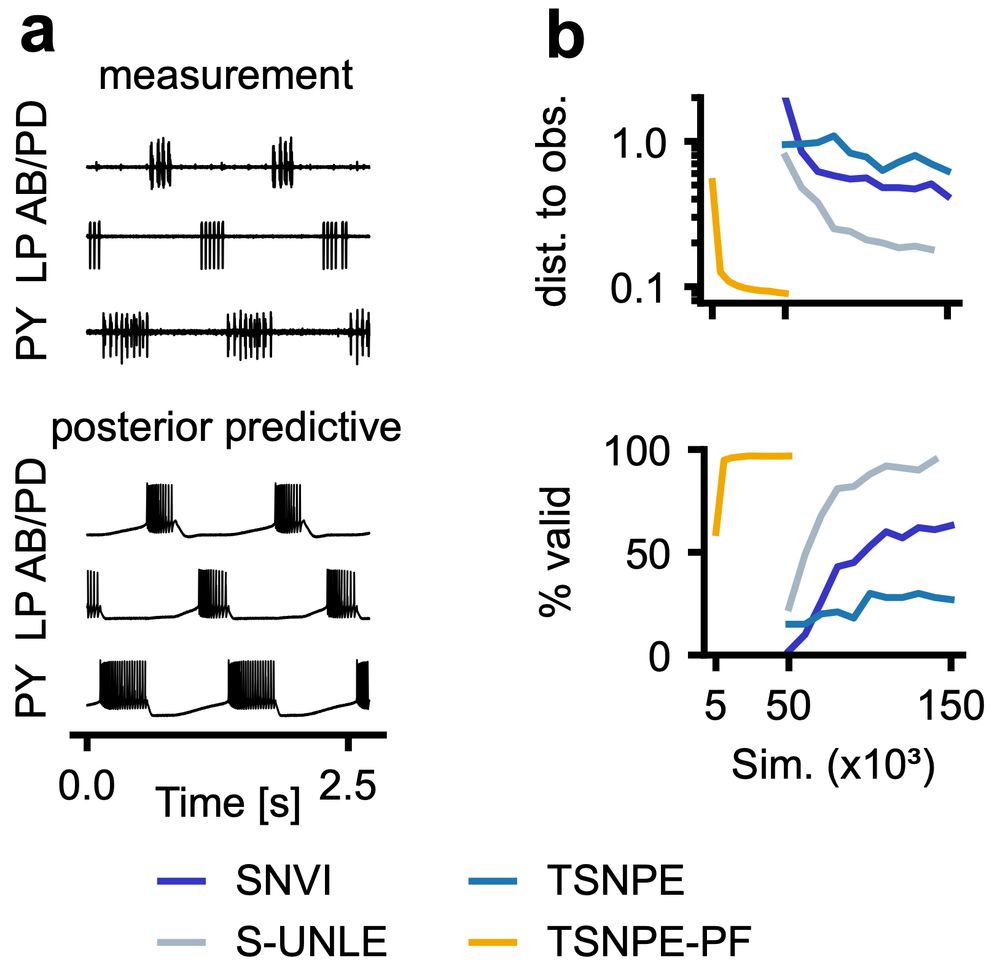
🧠 single-compartment neuron
🦀 31-parameter crab pyloric network
NPE-PF delivers tight posteriors & accurate predictions with far fewer simulations than previous methods.
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
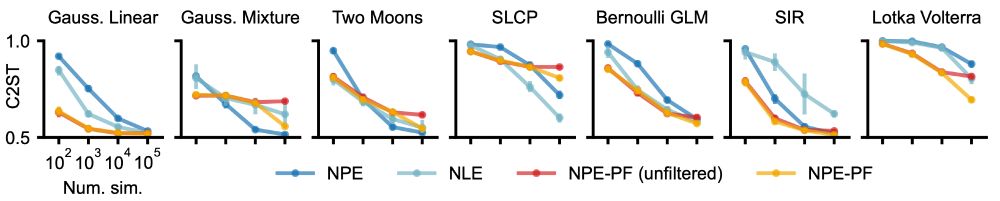
🚫 No need to train inference nets or tune hyperparameters.
🌟 Competitive or superior performance vs. standard SBI methods.
🚀 Especially strong performance for smaller simulation budgets.
🔄 Filtering to handle large datasets + support for sequential inference.
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
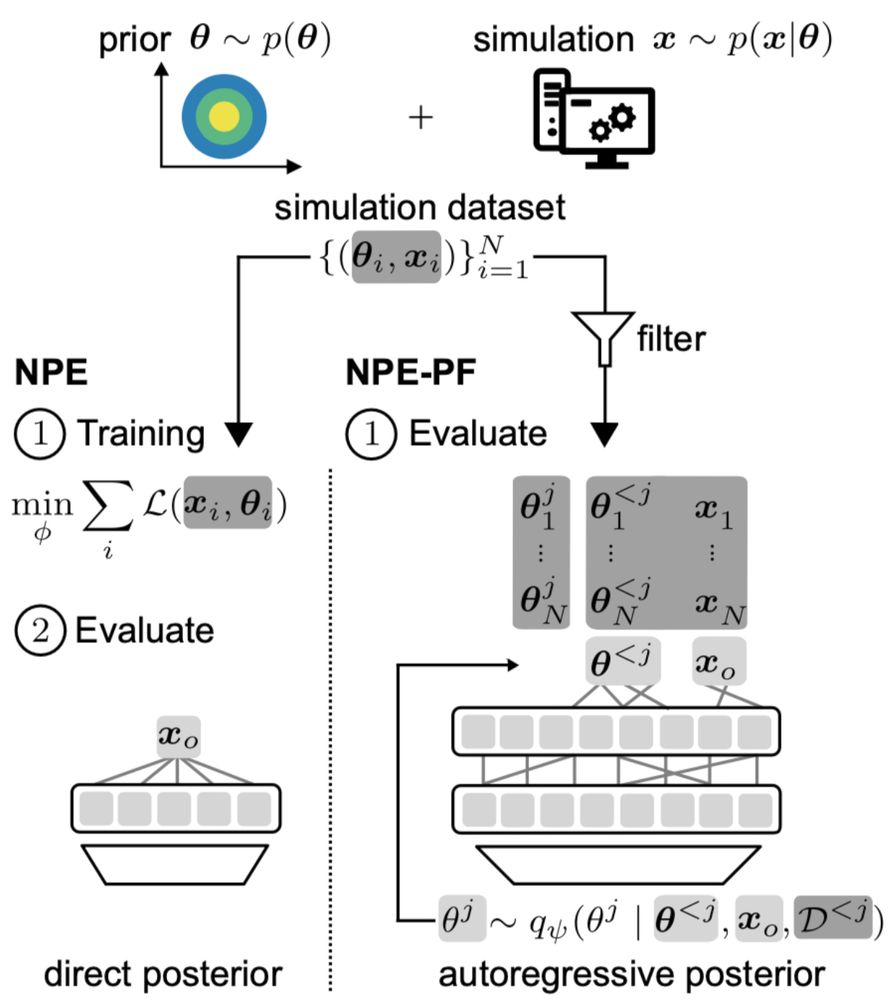
TabPFN, originally trained for tabular regression and classification, can estimate posteriors by autoregressively modeling one parameter dimension after the other.
It’s remarkably effective, even though TabPFN was not designed for SBI.
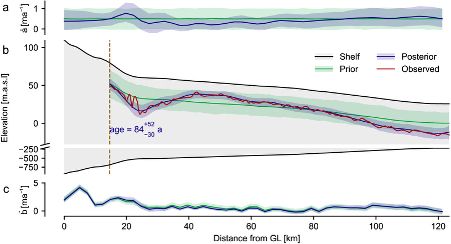
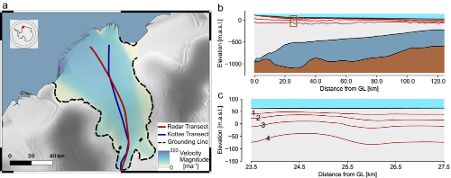
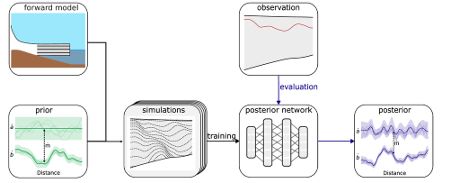
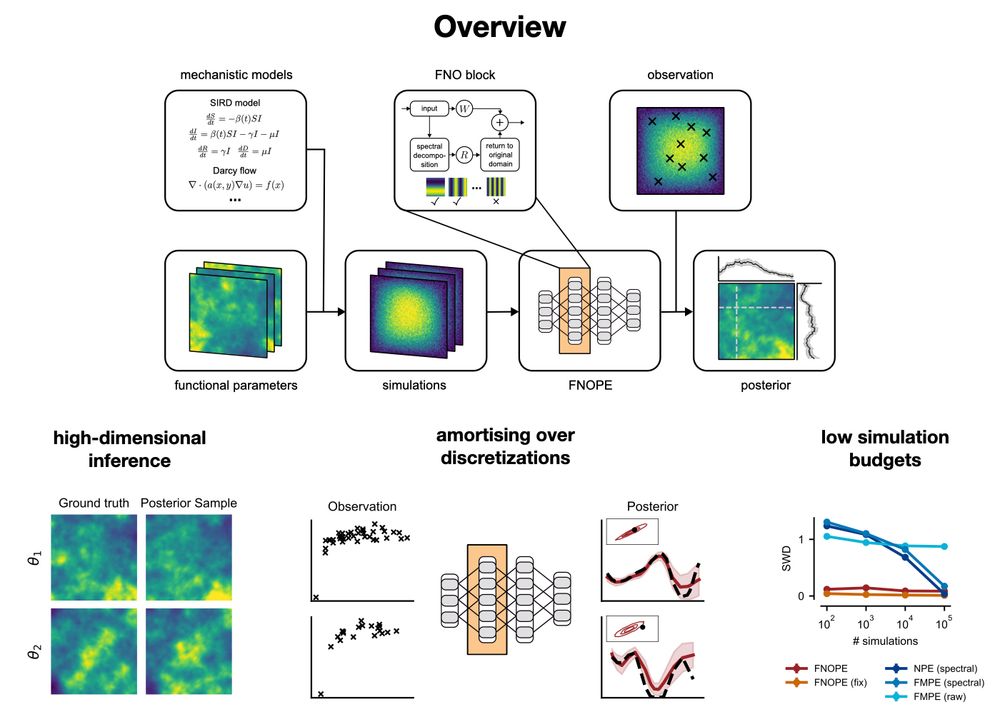
We validate this on a high-dimensional Kolmogorov flow simulator with around one million data dimensions.
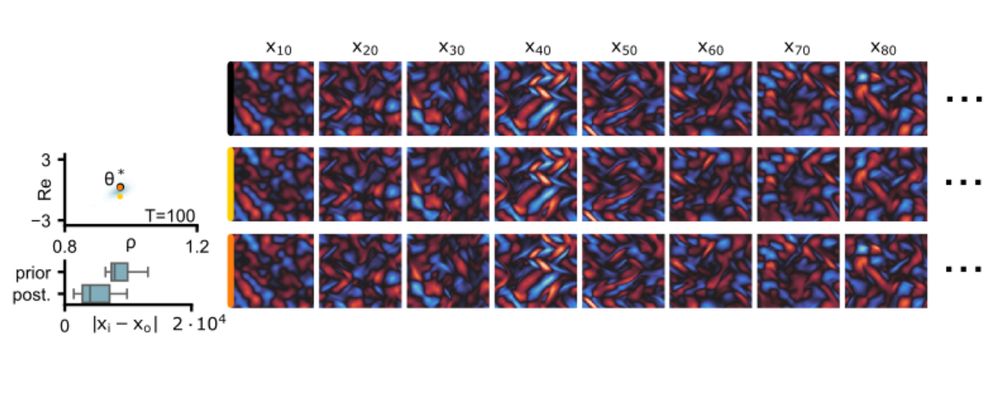
We validate this on a high-dimensional Kolmogorov flow simulator with around one million data dimensions.
Compared to NPE with embedding nets, it’s more simulation-efficient and accurate across time series of varying lengths.
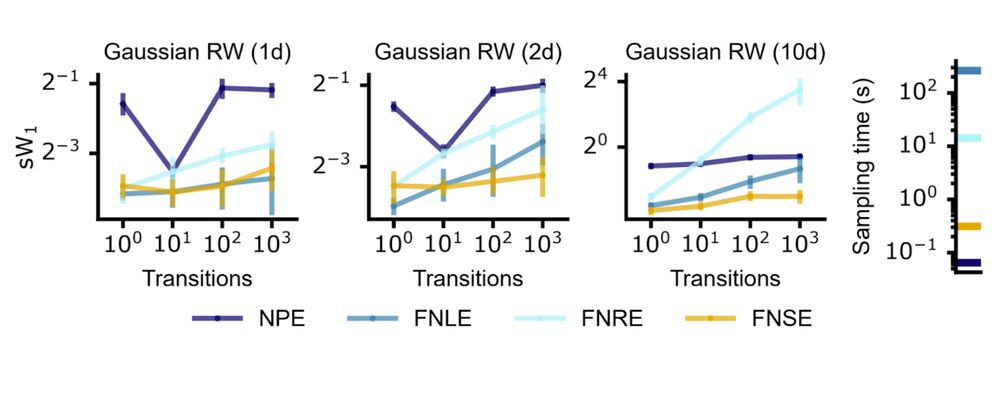
Compared to NPE with embedding nets, it’s more simulation-efficient and accurate across time series of varying lengths.
We then compose these local results to obtain a posterior over parameters that align with the entire time series observation.
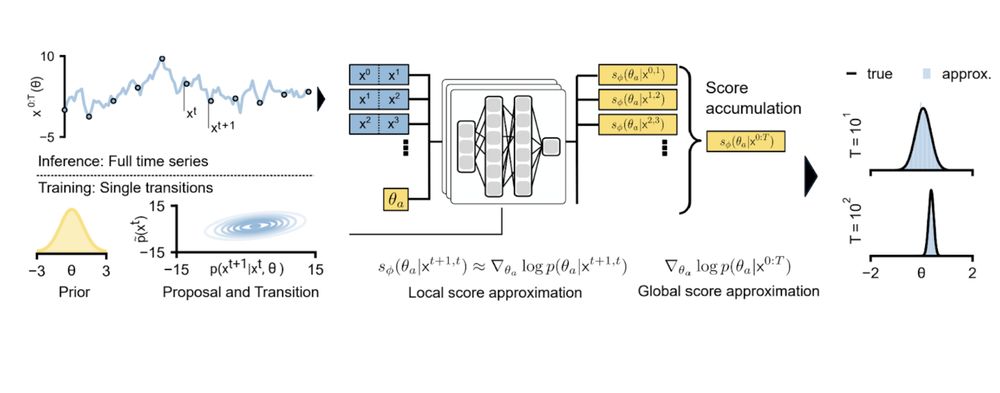
We then compose these local results to obtain a posterior over parameters that align with the entire time series observation.
Go to www.mackelab.org/jobs/ for details on more projects and how to contact us, or just find one of us (Auguste, Matthijs, Richard, Zina) at the conference—we're happy to chat!

Go to www.mackelab.org/jobs/ for details on more projects and how to contact us, or just find one of us (Auguste, Matthijs, Richard, Zina) at the conference—we're happy to chat!
He will tell you about the good, the bad, and the ugly of getting more than what you had asked for using mechanistic models and probabilistic machine learning.
17:00, Corriveau/Sateux

He will tell you about the good, the bad, and the ugly of getting more than what you had asked for using mechanistic models and probabilistic machine learning.
17:00, Corriveau/Sateux
Tuesday at 11:20, @auschulz.bsky.social will give a talk about deep generative models - VAEs and DDPMs - for linking neural activity and behavior, at the workshop on
"Building a foundation model for the brain" (Soutana 1).

Tuesday at 11:20, @auschulz.bsky.social will give a talk about deep generative models - VAEs and DDPMs - for linking neural activity and behavior, at the workshop on
"Building a foundation model for the brain" (Soutana 1).

