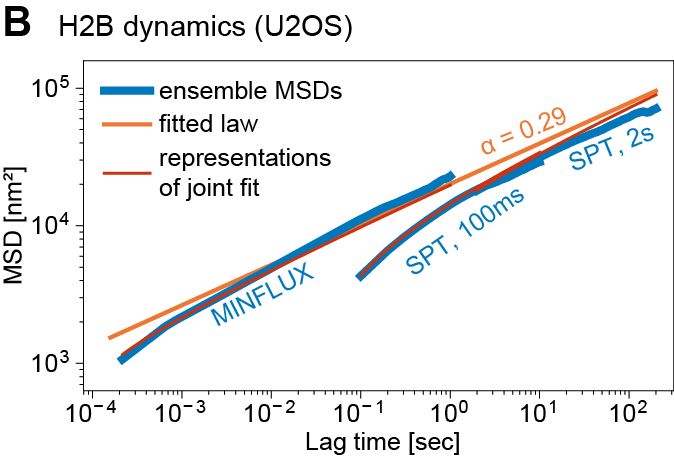
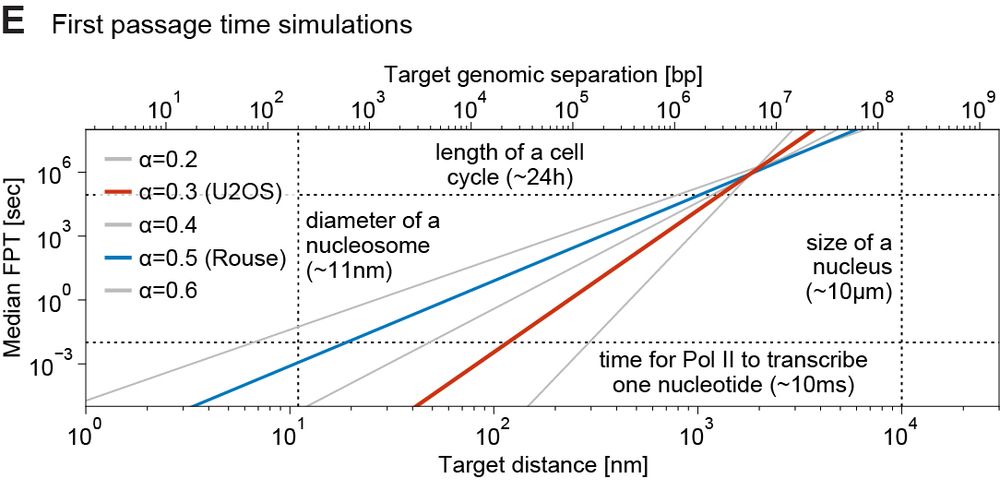
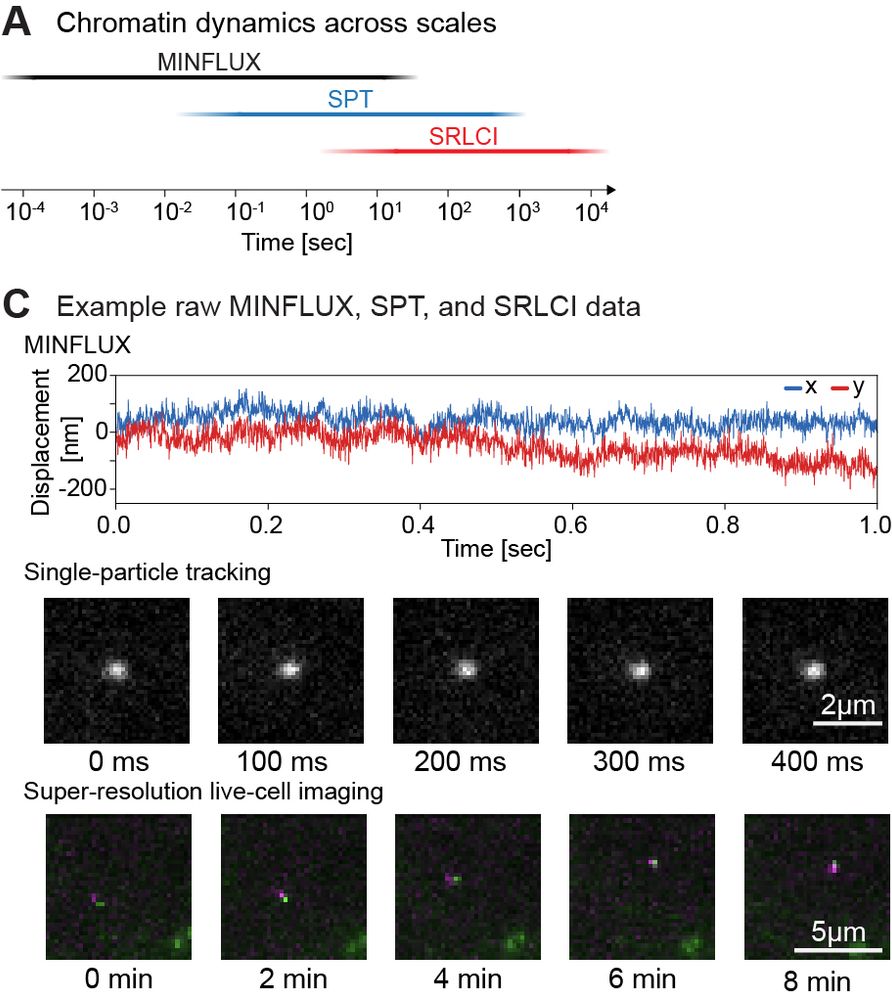
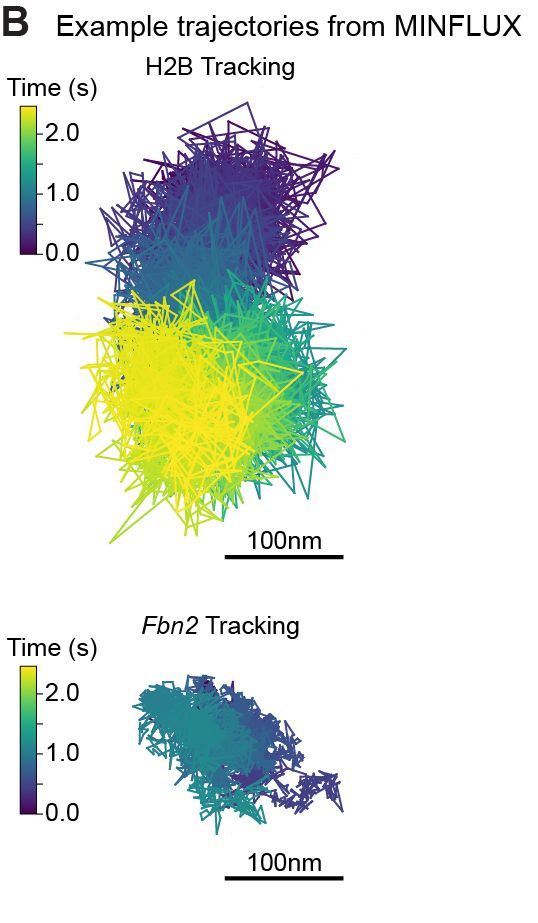
@matteomazzocca.bsky.social
60 followers
29 following
5 posts
Postdoc @andersshansen.bsky.social Lab @ MIT | PhD @davidemazza.bsky.social Lab @ Vita-Salute San Raffaele University | 3D genome, TFs, gene regulation
Posts
Media
Videos
Starter Packs